Antigen tests for COVID-19
- PMID: 33954080
- PMCID: PMC8049777
- DOI: 10.2142/biophysico.bppb-v18.004
Antigen tests for COVID-19
Abstract
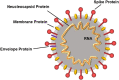
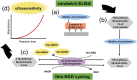
PCR diagnosis has been considered as the gold standard for coronavirus disease 2019 (COVID-19) and other many diseases. However, there are many problems in using PCR, such as non-specific (i.e., false-positive) and false-negative amplifications, the limits of a target sample volume, deactivation of the enzymes used, complicated techniques, difficulty in designing probe sequences, and the expense. We, thus, need an alternative to PCR, for example an ultrasensitive antigen test. In the present review, we summarize the following three topics. (1) The problems of PCR are outlined. (2) The antigen tests are surveyed in the literature that was published in 2020, and their pros and cons are discussed for commercially available antigen tests. (3) Our own antigen test on the basis of an ultrasensitive enzyme-linked immunosorbent assay (ELISA) is introduced. Finally, we discuss the possibility that our antigen test by an ultrasensitive ELISA technique will become the gold standard for diagnosis of COVID-19 and other diseases.
Keywords: COVID-19; SARS-CoV-2; antigen test; real-time PCR; ultrasensitive ELISA.
2021 THE BIOPHYSICAL SOCIETY OF JAPAN.
Figures

References
-
- Zhang Lab, Genome-wide structure and function modeling of SARS-CoV-2. Available online: https://zhanglab.ccmb.med.umich.edu/COVID-19/ (accessed on January 25, 2021).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

