The germline mutational process in rhesus macaque and its implications for phylogenetic dating
- PMID: 33954793
- PMCID: PMC8099771
- DOI: 10.1093/gigascience/giab029
The germline mutational process in rhesus macaque and its implications for phylogenetic dating
Abstract
Background: Understanding the rate and pattern of germline mutations is of fundamental importance for understanding evolutionary processes.
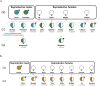
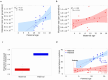
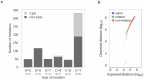
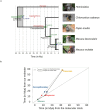
Results: Here we analyzed 19 parent-offspring trios of rhesus macaques (Macaca mulatta) at high sequencing coverage of ∼76× per individual and estimated a mean rate of 0.77 × 10-8de novo mutations per site per generation (95% CI: 0.69 × 10-8 to 0.85 × 10-8). By phasing 50% of the mutations to parental origins, we found that the mutation rate is positively correlated with the paternal age. The paternal lineage contributed a mean of 81% of the de novo mutations, with a trend of an increasing male contribution for older fathers. Approximately 3.5% of de novo mutations were shared between siblings, with no parental bias, suggesting that they arose from early development (postzygotic) stages. Finally, the divergence times between closely related primates calculated on the basis of the yearly mutation rate of rhesus macaque generally reconcile with divergence estimated with molecular clock methods, except for the Cercopithecoidea/Hominoidea molecular divergence dated at 58 Mya using our new estimate of the yearly mutation rate.
Conclusions: When compared to the traditional molecular clock methods, new estimated rates from pedigree samples can provide insights into the evolution of well-studied groups such as primates.
Keywords: Evolution; mutation rate; phylogeny; primates.
© The Author(s) 2021. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

