Transcriptional signatures of synaptic vesicle genes define myotonic dystrophy type I neurodegeneration
- PMID: 33955002
- PMCID: PMC9292638
- DOI: 10.1111/nan.12725
Transcriptional signatures of synaptic vesicle genes define myotonic dystrophy type I neurodegeneration
Abstract
Aim: To delineate the neurogenetic profiles of brain degeneration patterns in myotonic dystrophy type I (DM1).
Methods: In two cohorts of DM1 patients, brain maps of volume loss (VL) and neuropsychological deficits (NDs) were intersected to large-scale transcriptome maps provided by the Allen Human Brain Atlas (AHBA). For validation, neuropathological and RNA analyses were performed in a small series of DM1 brain samples.
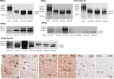
Results: Twofold: (1) From a list of preselected hypothesis-driven genes, confirmatory analyses found that three genes play a major role in brain degeneration: dystrophin (DMD), alpha-synuclein (SNCA) and the microtubule-associated protein tau (MAPT). Neuropathological analyses confirmed a highly heterogeneous Tau-pathology in DM1, different to the one in Alzheimer's disease. (2) Exploratory analyses revealed gene clusters enriched for key biological processes in the central nervous system, such as synaptic vesicle recycling, localization, endocytosis and exocytosis, and the serotonin and dopamine neurotransmitter pathways. RNA analyses confirmed synaptic vesicle dysfunction.
Conclusions: The combination of large-scale transcriptome interactions with brain imaging and cognitive function sheds light on the neurobiological mechanisms of brain degeneration in DM1 that might help define future therapeutic strategies and research into this condition.
Keywords: Allen Human Brain Atlas; DM1; neuropsychological deficits; structural neuroimaging; synaptic vesicles; volume loss.
© 2021 The Authors. Neuropathology and Applied Neurobiology published by John Wiley & Sons Ltd on behalf of British Neuropathological Society.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Lund M, Diaz LJ, Ranthe MF, et al. Cardiac involvement in myotonic dystrophy: a nationwide cohort study. Eur Heart J. 2014;35(32):2158‐2164. - PubMed
-
- Bogaard JM, van der Meché FG, Hendriks I, Ververs C. Pulmonary function and resting breathing pattern in myotonic dystrophy. Lung. 1992;170(3):143‐153. - PubMed
-
- Ørngreen MC, Arlien‐Søborg P, Duno M, Hertz JM, Vissing J. Endocrine function in 97 patients with myotonic dystrophy type 1. J Neurol. 2012;259(5):912‐920. - PubMed
-
- Laberge L, Gagnon C, Dauvilliers Y. Daytime sleepiness and myotonic dystrophy. Curr Neurol Neurosci Rep. 2013;13(4):340. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

