A 20-kb lineage-specific genomic region tames virulence in pathogenic amphidiploid Verticillium longisporum
- PMID: 33955130
- PMCID: PMC8295516
- DOI: 10.1111/mpp.13071
A 20-kb lineage-specific genomic region tames virulence in pathogenic amphidiploid Verticillium longisporum
Abstract
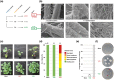
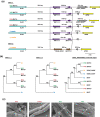
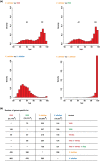
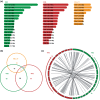
Amphidiploid fungal Verticillium longisporum strains Vl43 and Vl32 colonize the plant host Brassica napus but differ in their ability to cause disease symptoms. These strains represent two V. longisporum lineages derived from different hybridization events of haploid parental Verticillium strains. Vl32 and Vl43 carry same-sex mating-type genes derived from both parental lineages. Vl32 and Vl43 similarly colonize and penetrate plant roots, but asymptomatic Vl32 proliferation in planta is lower than virulent Vl43. The highly conserved Vl43 and Vl32 genomes include less than 1% unique genes, and the karyotypes of 15 or 16 chromosomes display changed genetic synteny due to substantial genomic reshuffling. A 20 kb Vl43 lineage-specific (LS) region apparently originating from the Verticillium dahliae-related ancestor is specific for symptomatic Vl43 and encodes seven genes, including two putative transcription factors. Either partial or complete deletion of this LS region in Vl43 did not reduce virulence but led to induction of even more severe disease symptoms in rapeseed. This suggests that the LS insertion in the genome of symptomatic V. longisporum Vl43 mediates virulence-reducing functions, limits damage on the host plant, and therefore tames Vl43 from being even more virulent.
Keywords: Verticillium longisporum; genome comparison; hybridization; lineage-specific region; pathogenicity.
© 2021 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures

References
-
- Almagro Armenteros, J.J. , Sønderby, C.K. , Sønderby, S.K. , Nielsen, H. & Winther, O. (2017) DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics, 33, 3387–3395. - PubMed
-
- Bui, T.‐T. , Harting, R. , Braus‐Stromeyer, S.A. , Tran, V.‐T. , Leonard, M. , Höfer, A. et al. (2019) Verticillium dahliae transcription factors Som1 and Vta3 control microsclerotia formation and sequential steps of plant root penetration and colonisation to induce disease. New Phytologist, 221, 2138–2159. - PubMed
-
- Caracuel, Z. , Roncero, M.I.G. , Espeso, E.A. , González‐Verdejo, C.I. , García‐Maceira, F.I. & Di Pietro, A. (2003) The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum . Molecular Microbiology, 48, 765–779. - PubMed
-
- Casadevall, A. & Pirofski, L.‐A. (2014) Microbiology: Ditch the term pathogen. Nature, 516, 165–166. - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

