How Protein Methylation Regulates Steroid Receptor Function
- PMID: 33955470
- PMCID: PMC8755998
- DOI: 10.1210/endrev/bnab014
How Protein Methylation Regulates Steroid Receptor Function
Abstract
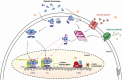
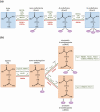
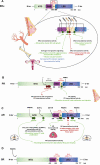
Steroid receptors (SRs) are members of the nuclear hormonal receptor family, many of which are transcription factors regulated by ligand binding. SRs regulate various human physiological functions essential for maintenance of vital biological pathways, including development, reproduction, and metabolic homeostasis. In addition, aberrant expression of SRs or dysregulation of their signaling has been observed in a wide variety of pathologies. SR activity is tightly and finely controlled by post-translational modifications (PTMs) targeting the receptors and/or their coregulators. Whereas major attention has been focused on phosphorylation, growing evidence shows that methylation is also an important regulator of SRs. Interestingly, the protein methyltransferases depositing methyl marks are involved in many functions, from development to adult life. They have also been associated with pathologies such as inflammation, as well as cardiovascular and neuronal disorders, and cancer. This article provides an overview of SR methylation/demethylation events, along with their functional effects and biological consequences. An in-depth understanding of the landscape of these methylation events could provide new information on SR regulation in physiology, as well as promising perspectives for the development of new therapeutic strategies, illustrated by the specific inhibitors of protein methyltransferases that are currently available.
Keywords: AR; ERα; GR; PR; coregulators; lysine methyltransferases; methylation; protein arginine methyltransferases; protein demethylases; steroid receptors.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Endocrine Society.
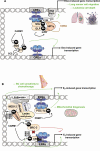
Figures

References
-
- Baschant U, Lane NE, Tuckermann J. The multiple facets of glucocorticoid action in rheumatoid arthritis. Nat Rev Rheumatol. 2012;8(11):645-655. - PubMed
-
- Ashburner M. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. II. The effects of inhibitors of protein synthesis. Dev Biol. 1974;39(1):141-157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

