UGT76B1, a promiscuous hub of small molecule-based immune signaling, glucosylates N-hydroxypipecolic acid, and balances plant immunity
- PMID: 33955482
- PMCID: PMC8136890
- DOI: 10.1093/plcell/koaa044
UGT76B1, a promiscuous hub of small molecule-based immune signaling, glucosylates N-hydroxypipecolic acid, and balances plant immunity
Abstract
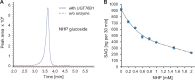
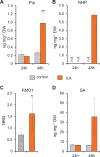
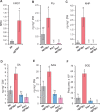
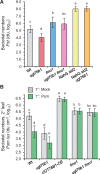
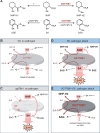
Glucosylation modulates the biological activity of small molecules and frequently leads to their inactivation. The Arabidopsis thaliana glucosyltransferase UGT76B1 is involved in conjugating the stress hormone salicylic acid (SA) as well as isoleucic acid (ILA). Here, we show that UGT76B1 also glucosylates N-hydroxypipecolic acid (NHP), which is synthesized by FLAVIN-DEPENDENT MONOOXYGENASE 1 (FMO1) and activates systemic acquired resistance (SAR). Upon pathogen attack, Arabidopsis leaves generate two distinct NHP hexose conjugates, NHP-O-β-glucoside and NHP glucose ester, whereupon only NHP-O-β-glucoside formation requires a functional SA pathway. The ugt76b1 mutants specifically fail to generate the NHP-O-β-glucoside, and recombinant UGT76B1 synthesizes NHP-O-β-glucoside in vitro in competition with SA and ILA. The loss of UGT76B1 elevates the endogenous levels of NHP, SA, and ILA and establishes a constitutive SAR-like immune status. Introgression of the fmo1 mutant lacking NHP biosynthesis into the ugt76b1 background abolishes this SAR-like resistance. Moreover, overexpression of UGT76B1 in Arabidopsis shifts the NHP and SA pools toward O-β-glucoside formation and abrogates pathogen-induced SAR. Our results further indicate that NHP-triggered immunity is SA-dependent and relies on UGT76B1 as a common metabolic hub. Thereby, UGT76B1-mediated glucosylation controls the levels of active NHP, SA, and ILA in concert to balance the plant immune status.
© The Author(s) 2021. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures



References
-
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid–independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 - PMC - PubMed
-
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

