A Hill type equation can predict target gene expression driven by p53 pulsing
- PMID: 33955710
- PMCID: PMC8167869
- DOI: 10.1002/2211-5463.13179
A Hill type equation can predict target gene expression driven by p53 pulsing
Erratum in
-
Corrigendum to: A Hill type equation can predict target gene expression driven by p53 pulsing. https://febs.onlinelibrary.wiley.com/doi/full/10.1002/2211-5463.13179.FEBS Open Bio. 2024 Jul;14(7):1218. doi: 10.1002/2211-5463.13838. Epub 2024 May 30. FEBS Open Bio. 2024. PMID: 38817047 Free PMC article. No abstract available.
Abstract
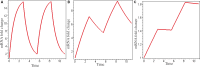
Many factors determine target gene expression dynamics under p53 pulsing. In this study, I sought to determine the mechanism by which duration, frequency, binding affinity and maximal transcription rate affect the expression dynamics of target genes. Using an analytical method to solve a simple model, I found that the fold change of target gene expression increases relative to the number of p53 pulses, and the optimal frequency, 0.18 h-1 , from two real p53 pulses drives the maximal fold change with a decay rate of 0.18 h-1 . Moreover, p53 pulses may also lead to a higher fold change than sustained p53. Finally, I discovered that a Hill-type equation, including these effect factors, can characterise target gene expression. The average error between the theoretical predictions and experiments was 23%. Collectively, this equation advances the understanding of transcription factor dynamics, where duration and frequency play a significant role in the fine regulation of target gene expression with higher binding affinity.
Keywords: fold change; hill equation; mRNA dynamical model; p53 pulse; target gene expression patterns.
© 2021 The Authors. FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest'.
Figures

Similar articles
-
p53 pulses lead to distinct patterns of gene expression albeit similar DNA-binding dynamics.Nat Struct Mol Biol. 2017 Oct;24(10):840-847. doi: 10.1038/nsmb.3452. Epub 2017 Aug 21. Nat Struct Mol Biol. 2017. PMID: 28825732 Free PMC article.
-
p53 Pulses Diversify Target Gene Expression Dynamics in an mRNA Half-Life-Dependent Manner and Delineate Co-regulated Target Gene Subnetworks.Cell Syst. 2016 Apr 27;2(4):272-82. doi: 10.1016/j.cels.2016.03.006. Epub 2016 Apr 7. Cell Syst. 2016. PMID: 27135539 Free PMC article.
-
Understanding non-linear effects from Hill-type dynamics with application to decoding of p53 signaling.Sci Rep. 2018 Feb 1;8(1):2147. doi: 10.1038/s41598-018-20466-2. Sci Rep. 2018. PMID: 29391550 Free PMC article.
-
Progress and challenges in understanding the regulation and function of p53 dynamics.Biochem Soc Trans. 2021 Nov 1;49(5):2123-2131. doi: 10.1042/BST20210148. Biochem Soc Trans. 2021. PMID: 34495325 Free PMC article. Review.
-
To repress or not to repress: this is the guardian's question.Trends Cell Biol. 2011 Jun;21(6):344-53. doi: 10.1016/j.tcb.2011.04.002. Epub 2011 May 19. Trends Cell Biol. 2011. PMID: 21601459 Review.
Cited by
-
The Hill-Type Equation Reveals the Regulatory Principle of Target Protein Expression Led by p53 Pulsing.FASEB Bioadv. 2025 Jun 6;7(8):e70026. doi: 10.1096/fba.2024-00220. eCollection 2025 Aug. FASEB Bioadv. 2025. PMID: 40746865 Free PMC article.
-
A New Model of Hemoglobin Oxygenation.Entropy (Basel). 2022 Aug 30;24(9):1214. doi: 10.3390/e24091214. Entropy (Basel). 2022. PMID: 36141103 Free PMC article.
References
-
- Levine AJ (2019) Targeting therapies for the p53 protein in cancer treatments. Annu Rev Cancer Biol 3, 21–34.
-
- Kolch W and Kiel C (2018) From oncogenic mutation to dynamic code. Science 361, 844–845. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

