From structure to the dynamic regulation of a molecular switch: A journey over 3 decades
- PMID: 33957122
- PMCID: PMC8144671
- DOI: 10.1016/j.jbc.2021.100746
From structure to the dynamic regulation of a molecular switch: A journey over 3 decades
Abstract
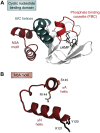
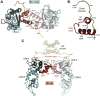
It is difficult to imagine where the signaling community would be today without the Protein Data Bank. This visionary resource, established in the 1970s, has been an essential partner for sharing information between academics and industry for over 3 decades. We describe here the history of our journey with the protein kinases using cAMP-dependent protein kinase as a prototype. We summarize what we have learned since the first structure, published in 1991, why our journey is still ongoing, and why it has been essential to share our structural information. For regulation of kinase activity, we focus on the cAMP-binding protein kinase regulatory subunits. By exploring full-length macromolecular complexes, we discovered not only allostery but also an essential motif originally attributed to crystal packing. Massive genomic data on disease mutations allows us to now revisit crystal packing as a treasure chest of possible protein:protein interfaces where the biological significance and disease relevance can be validated. It provides a new window into exploring dynamic intrinsically disordered regions that previously were deleted, ignored, or attributed to crystal packing. Merging of crystallography with cryo-electron microscopy, cryo-electron tomography, NMR, and millisecond molecular dynamics simulations is opening a new world for the signaling community where those structure coordinates, deposited in the Protein Data Bank, are just a starting point!
Keywords: allostery; cAMP; cAMP-dependent protein kinase (PKA); catalytic subunit; crystallography; dynamics; intrinsically disordered regions; protein kinases; protein structure; regulatory subunit.
Copyright © 2021. Published by Elsevier Inc.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

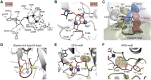





References
-
- Crystallography: Protein Data Bank. Nat. New Biol. 1971;233:223. - PubMed
-
- Knighton D.R., Zheng J.H., Ten Eyck L.F., Ashford V.A., Xuong N.H., Taylor S.S., Sowadski J.M. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. - PubMed
-
- Knighton D.R., Zheng J.H., Ten Eyck L.F., Xuong N.H., Taylor S.S., Sowadski J.M. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. - PubMed
-
- Zheng J., Knighton D.R., Ten Eyck L.F., Karlsson R., Xuong N., Taylor S.S., Sowadski J.M. Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry. 1993;32:2154–2161. - PubMed
-
- Ramakrishnan C., Dani V.S., Ramasarma T. A conformational analysis of Walker motif A [GXXXXGKT (S)] in nucleotide-binding and other proteins. Protein Eng. 2002;15:783–798. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

