Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11
- PMID: 33958322
- PMCID: PMC8172134
- DOI: 10.1126/sciadv.abf5632
Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11
Abstract
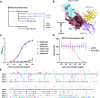
The emergence of SARS-CoV-2 antibody escape mutations highlights the urgent need for broadly neutralizing therapeutics. We previously identified a human monoclonal antibody, 47D11, capable of cross-neutralizing SARS-CoV-2 and SARS-CoV and protecting against the associated respiratory disease in an animal model. Here, we report cryo-EM structures of both trimeric spike ectodomains in complex with the 47D11 Fab. 47D11 binds to the closed receptor-binding domain, distal to the ACE2 binding site. The CDRL3 stabilizes the N343 glycan in an upright conformation, exposing a mutationally constrained hydrophobic pocket, into which the CDRH3 loop inserts two aromatic residues. 47D11 stabilizes a partially open conformation of the SARS-CoV-2 spike, suggesting that it could be used effectively in combination with other antibodies targeting the exposed receptor-binding motif. Together, these results reveal a cross-protective epitope on the SARS-CoV-2 spike and provide a structural roadmap for the development of 47D11 as a prophylactic or postexposure therapy for COVID-19.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution License 4.0 (CC BY).
Figures



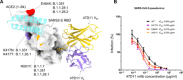
References
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). - PMC - PubMed
-
- Ksiazek T. G., Erdman D., Goldsmith C. S., Zaki S. R., Peret T., Emery S., Tong S., Urbani C., Comer J. A., Lim W., Rollin P. E., Dowell S. F., Ling A.-E., Humphrey C. D., Shieh W.-J., Guarner J., Paddock C. D., Rota P., Fields B., DeRisi J., Yang J.-Y., Cox N., Hughes J. M., LeDuc J. W., Bellini W. J., Anderson L. J.; SARS Working Group , A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

