Identification of germ cell-specific Mga variant mRNA that promotes meiosis via impediment of a non-canonical PRC1
- PMID: 33958653
- PMCID: PMC8102552
- DOI: 10.1038/s41598-021-89123-5
Identification of germ cell-specific Mga variant mRNA that promotes meiosis via impediment of a non-canonical PRC1
Abstract
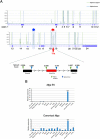
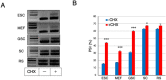
A non-canonical PRC1 (PRC1.6) prevents precocious meiotic onset. Germ cells alleviate its negative effect by reducing their amount of MAX, a component of PRC1.6, as a prerequisite for their bona fide meiosis. Here, we found that germ cells produced Mga variant mRNA bearing a premature termination codon (PTC) during meiosis as an additional mechanism to impede the function of PRC1.6. The variant mRNA encodes an anomalous MGA protein that lacks the bHLHZ domain and thus functions as a dominant negative regulator of PRC1.6. Notwithstanding the presence of PTC, the Mga variant mRNA are rather stably present in spermatocytes and spermatids due to their intrinsic inefficient background of nonsense-mediated mRNA decay. Thus, our data indicate that meiosis is controlled in a multi-layered manner in which both MAX and MGA, which constitute the core of PRC1.6, are at least used as targets to deteriorate the integrity of the complex to ensure progression of meiosis.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Alternative splicing for germ cell-specific Mga transcript can be eliminated without compromising mouse viability or fertility.Dev Growth Differ. 2022 Sep;64(7):409-416. doi: 10.1111/dgd.12806. Epub 2022 Sep 14. Dev Growth Differ. 2022. PMID: 36053973
-
MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6.PLoS Genet. 2018 Jan 30;14(1):e1007193. doi: 10.1371/journal.pgen.1007193. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29381691 Free PMC article.
-
Two DNA binding domains of MGA act in combination to suppress ectopic activation of meiosis-related genes in mouse embryonic stem cells.Stem Cells. 2021 Nov;39(11):1435-1446. doi: 10.1002/stem.3433. Epub 2021 Jul 14. Stem Cells. 2021. PMID: 34224650
-
[Controlling the spatiotemporal expression of germ line specific genes by PRC1.6 complex].Yi Chuan. 2019 Apr 20;41(4):271-284. doi: 10.16288/j.yczz.18-332. Yi Chuan. 2019. PMID: 30992249 Review. Chinese.
-
Does MAX open up a new avenue for meiotic research?Dev Growth Differ. 2017 Feb;59(2):61-69. doi: 10.1111/dgd.12344. Epub 2017 Feb 20. Dev Growth Differ. 2017. PMID: 28220481 Review.
Cited by
-
SMG6 localizes to the chromatoid body and shapes the male germ cell transcriptome to drive spermatogenesis.Nucleic Acids Res. 2022 Nov 11;50(20):11470-11491. doi: 10.1093/nar/gkac900. Nucleic Acids Res. 2022. PMID: 36259644 Free PMC article.
-
Destabilization of mRNAs enhances competence to initiate meiosis in mouse spermatogenic cells.Development. 2024 Jul 15;151(14):dev202740. doi: 10.1242/dev.202740. Epub 2024 Jul 15. Development. 2024. PMID: 38884383 Free PMC article.
-
PFOA-Induced Ovotoxicity Differs Between Lean and Obese Mice With Impacts on Ovarian Reproductive and DNA Damage Sensing and Repair Proteins.Toxicol Sci. 2022 Nov 23;190(2):173-188. doi: 10.1093/toxsci/kfac104. Toxicol Sci. 2022. PMID: 36214631 Free PMC article.
-
Loss of MGA repression mediated by an atypical polycomb complex promotes tumor progression and invasiveness.Elife. 2021 Jul 8;10:e64212. doi: 10.7554/eLife.64212. Elife. 2021. PMID: 34236315 Free PMC article.
-
Repression of germline genes by PRC1.6 and SETDB1 in the early embryo precedes DNA methylation-mediated silencing.Nat Commun. 2021 Dec 2;12(1):7020. doi: 10.1038/s41467-021-27345-x. Nat Commun. 2021. PMID: 34857746 Free PMC article.
References
-
- Ginsburg M, Snow MH, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. - PubMed
-
- Ishiguro KI, et al. MEIOSIN directs the switch from mitosis to meiosis in mammalian germ cells. Dev. Cell. 2020;52:429–445. - PubMed
-
- Yokobayashi S, et al. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature. 2013;495:236–240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

