LIMIT is an immunogenic lncRNA in cancer immunity and immunotherapy
- PMID: 33958760
- PMCID: PMC8122078
- DOI: 10.1038/s41556-021-00672-3
LIMIT is an immunogenic lncRNA in cancer immunity and immunotherapy
Abstract
Major histocompatibility complex-I (MHC-I) presents tumour antigens to CD8+ T cells and triggers anti-tumour immunity. Humans may have 30,000-60,000 long noncoding RNAs (lncRNAs). However, it remains poorly understood whether lncRNAs affect tumour immunity. Here, we identify a lncRNA, lncRNA inducing MHC-I and immunogenicity of tumour (LIMIT), in humans and mice. We found that IFNγ stimulated LIMIT, LIMIT cis-activated the guanylate-binding protein (GBP) gene cluster and GBPs disrupted the association between HSP90 and heat shock factor-1 (HSF1), thereby resulting in HSF1 activation and transcription of MHC-I machinery, but not PD-L1. RNA-guided CRISPR activation of LIMIT boosted GBPs and MHC-I, and potentiated tumour immunogenicity and checkpoint therapy. Silencing LIMIT, GBPs and/or HSF1 diminished MHC-I, impaired antitumour immunity and blunted immunotherapy efficacy. Clinically, LIMIT, GBP- and HSF1-signalling transcripts and proteins correlated with MHC-I, tumour-infiltrating T cells and checkpoint blockade response in patients with cancer. Together, we demonstrate that LIMIT is a cancer immunogenic lncRNA and the LIMIT-GBP-HSF1 axis may be targetable for cancer immunotherapy.
Conflict of interest statement
Competing Interest Declaration
W.Z. has served as a scientific advisor or consultant for NGM, Cstone, Oncopia, and Hengenix. All other authors declare no competing interests.
Figures
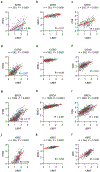




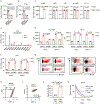









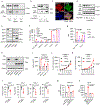
Comment in
-
LIMITing tumours with an immunogenic lncRNA.Nat Cell Biol. 2021 May;23(5):443-445. doi: 10.1038/s41556-021-00682-1. Nat Cell Biol. 2021. PMID: 33958759 No abstract available.
Similar articles
-
RIG-I is an intracellular checkpoint that limits CD8+ T-cell antitumour immunity.EMBO Mol Med. 2024 Nov;16(11):3005-3025. doi: 10.1038/s44321-024-00136-9. Epub 2024 Sep 25. EMBO Mol Med. 2024. PMID: 39322862 Free PMC article.
-
The long noncoding RNA NEAT1 and nuclear paraspeckles are up-regulated by the transcription factor HSF1 in the heat shock response.J Biol Chem. 2018 Dec 7;293(49):18965-18976. doi: 10.1074/jbc.RA118.004473. Epub 2018 Oct 10. J Biol Chem. 2018. PMID: 30305397 Free PMC article.
-
MHC-II neoantigens shape tumour immunity and response to immunotherapy.Nature. 2019 Oct;574(7780):696-701. doi: 10.1038/s41586-019-1671-8. Epub 2019 Oct 23. Nature. 2019. PMID: 31645760 Free PMC article.
-
Recent Advances in Lung Cancer Immunotherapy: Input of T-Cell Epitopes Associated With Impaired Peptide Processing.Front Immunol. 2019 Jul 3;10:1505. doi: 10.3389/fimmu.2019.01505. eCollection 2019. Front Immunol. 2019. PMID: 31333652 Free PMC article. Review.
-
PD-1/PD-L1 axis regulation in cancer therapy: The role of long non-coding RNAs and microRNAs.Life Sci. 2020 Sep 1;256:117899. doi: 10.1016/j.lfs.2020.117899. Epub 2020 Jun 3. Life Sci. 2020. PMID: 32504749 Review.
Cited by
-
Potential role of long noncoding RNA RP5-881L22.5 as a novel biomarker and therapeutic target of colorectal cancer.World J Gastrointest Oncol. 2022 Nov 15;14(11):2108-2121. doi: 10.4251/wjgo.v14.i11.2108. World J Gastrointest Oncol. 2022. PMID: 36438707 Free PMC article.
-
Bioinformatics and Experimental Study Revealed LINC00982/ miR-183-5p/ABCA8 Axis Suppresses LUAD Progression.Curr Cancer Drug Targets. 2024;24(6):654-667. doi: 10.2174/0115680096266700231107071222. Curr Cancer Drug Targets. 2024. PMID: 38419344
-
Epigenetic Mechanisms Underlying Melanoma Resistance to Immune and Targeted Therapies.Cancers (Basel). 2022 Nov 28;14(23):5858. doi: 10.3390/cancers14235858. Cancers (Basel). 2022. PMID: 36497341 Free PMC article. Review.
-
Boosting Antitumor Immunity with an Expanded Neoepitope Landscape.Cancer Res. 2022 Oct 17;82(20):3637-3649. doi: 10.1158/0008-5472.CAN-22-1525. Cancer Res. 2022. PMID: 35904353 Free PMC article. Review.
-
Current Research Progress of the Role of LncRNA LEF1-AS1 in a Variety of Tumors.Front Cell Dev Biol. 2021 Dec 20;9:750084. doi: 10.3389/fcell.2021.750084. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34988073 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA046592/CA/NCI NIH HHS/United States
- R01 CA217510/CA/NCI NIH HHS/United States
- R01 CA120458/CA/NCI NIH HHS/United States
- R01 CA152470/CA/NCI NIH HHS/United States
- R01 CA213566/CA/NCI NIH HHS/United States
- R01 CA123088/CA/NCI NIH HHS/United States
- U01 CA120458/CA/NCI NIH HHS/United States
- R01 CA099985/CA/NCI NIH HHS/United States
- T32 CA009672/CA/NCI NIH HHS/United States
- R01 CA244827/CA/NCI NIH HHS/United States
- R01 CA193136/CA/NCI NIH HHS/United States
- R01 CA248430/CA/NCI NIH HHS/United States
- P50 CA186786/CA/NCI NIH HHS/United States
- R01 CA216919/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

