Genome-wide survival study identifies a novel synaptic locus and polygenic score for cognitive progression in Parkinson's disease
- PMID: 33958783
- PMCID: PMC8459648
- DOI: 10.1038/s41588-021-00847-6
Genome-wide survival study identifies a novel synaptic locus and polygenic score for cognitive progression in Parkinson's disease
Abstract
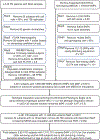
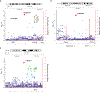
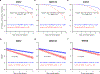
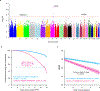
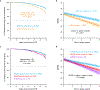
A key driver of patients' well-being and clinical trials for Parkinson's disease (PD) is the course that the disease takes over time (progression and prognosis). To assess how genetic variation influences the progression of PD over time to dementia, a major determinant for quality of life, we performed a longitudinal genome-wide survival study of 11.2 million variants in 3,821 patients with PD over 31,053 visits. We discover RIMS2 as a progression locus and confirm this in a replicate population (hazard ratio (HR) = 4.77, P = 2.78 × 10-11), identify suggestive evidence for TMEM108 (HR = 2.86, P = 2.09 × 10-8) and WWOX (HR = 2.12, P = 2.37 × 10-8) as progression loci, and confirm associations for GBA (HR = 1.93, P = 0.0002) and APOE (HR = 1.48, P = 0.001). Polygenic progression scores exhibit a substantial aggregate association with dementia risk, while polygenic susceptibility scores are not predictive. This study identifies a novel synaptic locus and polygenic score for cognitive disease progression in PD and proposes diverging genetic architectures of progression and susceptibility.
Figures

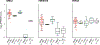
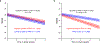
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

