Neuronal ApoE upregulates MHC-I expression to drive selective neurodegeneration in Alzheimer's disease
- PMID: 33958804
- PMCID: PMC9145692
- DOI: 10.1038/s41593-021-00851-3
Neuronal ApoE upregulates MHC-I expression to drive selective neurodegeneration in Alzheimer's disease
Abstract
Selective neurodegeneration is a critical causal factor in Alzheimer's disease (AD); however, the mechanisms that lead some neurons to perish, whereas others remain resilient, are unknown. We sought potential drivers of this selective vulnerability using single-nucleus RNA sequencing and discovered that ApoE expression level is a substantial driver of neuronal variability. Strikingly, neuronal expression of ApoE-which has a robust genetic linkage to AD-correlated strongly, on a cell-by-cell basis, with immune response pathways in neurons in the brains of wild-type mice, human ApoE knock-in mice and humans with or without AD. Elimination or over-expression of neuronal ApoE revealed a causal relationship among ApoE expression, neuronal MHC-I expression, tau pathology and neurodegeneration. Functional reduction of MHC-I ameliorated tau pathology in ApoE4-expressing primary neurons and in mouse hippocampi expressing pathological tau. These findings suggest a mechanism linking neuronal ApoE expression to MHC-I expression and, subsequently, to tau pathology and selective neurodegeneration.
Conflict of interest statement
Competing Interests Statement
Y.H. is a co-founder and scientific advisory board member of E-Scape Bio, Inc., GABAeron, Inc., and Mederon Bio, LLC. Other authors declare no competing financial interests.
Figures









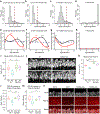
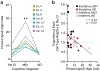

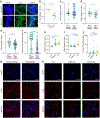
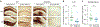
Comment in
-
Killing ageing neurons, one cell at a time.Nat Neurosci. 2021 Jun;24(6):759-760. doi: 10.1038/s41593-021-00855-z. Nat Neurosci. 2021. PMID: 33958803 No abstract available.
References
-
- Farrer LA et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: A meta-analysis. JAMA 278, 1349–1356 (1997). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

