Cell-Type-Specific Gene Modules Related to the Regional Homogeneity of Spontaneous Brain Activity and Their Associations With Common Brain Disorders
- PMID: 33958982
- PMCID: PMC8093778
- DOI: 10.3389/fnins.2021.639527
Cell-Type-Specific Gene Modules Related to the Regional Homogeneity of Spontaneous Brain Activity and Their Associations With Common Brain Disorders
Abstract
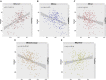
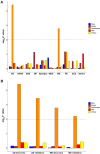
Mapping gene expression profiles to neuroimaging phenotypes in the same anatomical space provides opportunities to discover molecular substrates for human brain functional properties. Here, we aimed to identify cell-type-specific gene modules associated with the regional homogeneity (ReHo) of spontaneous brain activity and their associations with brain disorders. Fourteen gene modules were consistently associated with ReHo in the three datasets, five of which showed cell-type-specific expression (one neuron-endothelial module, one neuron module, one astrocyte module and two microglial modules) in two independent cell series of the human cerebral cortex. The neuron-endothelial module was mainly enriched for transporter complexes, the neuron module for the synaptic membrane, the astrocyte module for amino acid metabolism, and microglial modules for leukocyte activation and ribose phosphate biosynthesis. In enrichment analyses of cell-type-specific modules for 10 common brain disorders, only the microglial module was significantly enriched for genes obtained from genome-wide association studies of multiple sclerosis (MS) and Alzheimer's disease (AD). The ReHo of spontaneous brain activity is associated with the gene expression profiles of neurons, astrocytes, microglia and endothelial cells. The microglia-related genes associated with MS and AD may provide possible molecular substrates for ReHo abnormality in both brain disorders.
Keywords: Allen Human Brain Atlas; cell type; fMRI; gene expression; regional homogeneity.
Copyright © 2021 Shen, Yang, Xie, Wu, Zheng, Wang, Wang, Zhang, Li, Ye and Yu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

