Exploring the Antibiotic Resistance Burden in Livestock, Livestock Handlers and Their Non-Livestock Handling Contacts: A One Health Perspective
- PMID: 33959112
- PMCID: PMC8093850
- DOI: 10.3389/fmicb.2021.651461
Exploring the Antibiotic Resistance Burden in Livestock, Livestock Handlers and Their Non-Livestock Handling Contacts: A One Health Perspective
Abstract
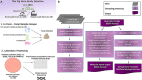
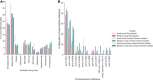
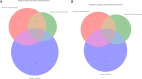
Antibiotics are freqeuently used in the livestock sector in low- and middle-income countries for treatment, prophylaxis, and growth promotion. However, there is limited information into the zoonotic prevalence and dissemination patterns of antimicrobial resistance (AMR) within these environments. In this study we used pig farming in Thailand as a model to explore AMR; 156 pig farms were included, comprising of small-sized (<50 sows) and medium-sized (≥100 sows) farms, where bacterial isolates were selectively cultured from animal rectal and human fecal samples. Bacterial isolates were subjected to antimicrobial susceptibility testing (AST), and whole-genome sequencing. Our results indicate extensive zoonotic sharing of antibiotic resistance genes (ARGs) by horizontal gene transfer. Resistance to multiple antibiotics was observed with higher prevalence in medium-scale farms. Zoonotic transmission of colistin resistance in small-scale farms had a dissemination gradient from pigs to handlers to non-livestock contacts. We highly recommend reducing the antimicrobial use in animals' feeds and medications, especially the last resort drug colistin.
Keywords: E. coli; antibiotic resistance; livestock; meat-production; one-health approach; pigs; zoonotic transmission.
Copyright © 2021 Hickman, Leangapichart, Lunha, Jiwakanon, Angkititrakul, Magnusson, Sunde and Järhult.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- 8.3rc1 Documentation (2020). 3.8.3rc1 Documentation. Delaware: Python. Available online at: https://docs.python.org/3/ (accessed May 12, 2020).
-
- Abdelgader S. A., Shi D., Chen M., Zhang L., Hejair H. M. A., Muhammad U., et al. (2018). Antibiotics Resistance Genes Screening and Comparative Genomics Analysis of Commensal Escherichia coli Isolated from Poultry Farms between China and Sudan. Biomed. Res. Int. 2018:5327450. 10.1155/2018/5327450 - DOI - PMC - PubMed
-
- Atterby C., Osbjer K., Tepper V., Rajala E., Hernandez J., Seng S., et al. (2019). Carriage of carbapenemase− and extended−spectrum cephalosporinase−producing Escherichia coli and Klebsiella pneumoniae in humans and livestock in rural Cambodia; gender and age differences and detection of bla OXA–48 in humans. Zoonoses Public Health 66 603–617. 10.1111/zph.12612 - DOI - PMC - PubMed
-
- Babraham Bioinformatics (2020). Trim Galore. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed May 12, 2020)
-
- Boehmer T., Vogler A. J., Thomas A., Sauer S., Hergenroether M., Straubinger R. K., et al. (2018). Phenotypic characterization and whole genome analysis of extended-spectrum beta-lactamase-producing bacteria isolated from dogs in Germany. PLoS One 13:e0206252. 10.1371/journal.pone.0206252 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

