Reck-Notch1 Signaling Mediates miR-221/222 Regulation of Lung Cancer Stem Cells in NSCLC
- PMID: 33959615
- PMCID: PMC8093830
- DOI: 10.3389/fcell.2021.663279
Reck-Notch1 Signaling Mediates miR-221/222 Regulation of Lung Cancer Stem Cells in NSCLC
Abstract
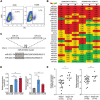
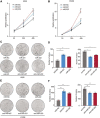
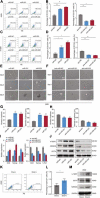
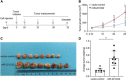
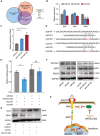
Cancer stem cells (CSCs) contribute to the cancer initiation, metastasis and drug resistance in non-small cell lung cancer (NSCLC). Herein, we identified a miR-221/222 cluster as a novel regulator of CSCs in NSCLC. Targeted overexpression or knockdown of miR-221/222 in NSCLC cells revealed the essential roles of miR-221/222 in regulation of lung cancer cell proliferation, mammosphere formation, subpopulation of CD133+ CSCs and the expression of stemness genes including OCT4, NANOG and h-TERT. The in vivo animal study showed that overexpression of miR-221/222 significantly enhanced the capacity of lung cancer cells to develop tumor and grow faster, indicating the importance of miR-221/222 in tumorigenesis and tumor growth. Mechanistically, Reck was found to be a key direct target gene of miR-221/222 in NSCLC. Overexpression of miR-221/222 significantly suppressed Reck expression, activated Notch1 signaling and increased the level of NICD. As an activated form of Notch1, NICD leads to enhanced stemness in NSCLC cells. In addition, knockdown of Reck by siRNA not only mimicked miR-221/222 effects, but also demonstrated involvement of Reck in the miR-221/222-induced activation of Notch1 signaling, verifying the essential roles of the miR-221/222-Reck-Notch1 axis in regulating stemness of NSCLC cells. These findings uncover a novel mechanism by which lung CSCs are significantly manipulated by miR-221/222, and provide a potential therapeutic target for the treatment of NSCLC.
Keywords: Notch1 signaling; Reck; cancer stem cell; miR-221/222; non-small cell lung cancer.
Copyright © 2021 Guo, Wang, Wang, Ding, Qian, Li, Ren, Liu, Ma, Li, Li, Zhao, Lü, Li, Wang and Yu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
RECK inhibits stemness gene expression and tumorigenicity of gastric cancer cells by suppressing ADAM-mediated Notch1 activation.J Cell Physiol. 2014 Feb;229(2):191-201. doi: 10.1002/jcp.24434. J Cell Physiol. 2014. PMID: 23881612
-
Long noncoding RNA MALAT1 mediates stem cell-like properties in human colorectal cancer cells by regulating miR-20b-5p/Oct4 axis.J Cell Physiol. 2019 Nov;234(11):20816-20828. doi: 10.1002/jcp.28687. Epub 2019 Apr 22. J Cell Physiol. 2019. PMID: 31012108
-
Intracellular Notch1 Signaling in Cancer-Associated Fibroblasts Dictates the Plasticity and Stemness of Melanoma Stem/Initiating Cells.Stem Cells. 2019 Jul;37(7):865-875. doi: 10.1002/stem.3013. Epub 2019 Apr 19. Stem Cells. 2019. PMID: 30941836 Free PMC article.
-
Suppression of Non-Small Cell Lung Cancer Growth and Metastasis by a Novel Small Molecular Activator of RECK.Cell Physiol Biochem. 2018;45(5):1807-1817. doi: 10.1159/000487872. Epub 2018 Feb 28. Cell Physiol Biochem. 2018. PMID: 29510387
-
miR-150-5p Inhibits Non-Small-Cell Lung Cancer Metastasis and Recurrence by Targeting HMGA2 and β-Catenin Signaling.Mol Ther Nucleic Acids. 2019 Jun 7;16:675-685. doi: 10.1016/j.omtn.2019.04.017. Epub 2019 Apr 23. Mol Ther Nucleic Acids. 2019. PMID: 31121479 Free PMC article.
Cited by
-
Tumor-suppressive E3 ubiquitin ligase CHIP inhibits the PBK/ERK axis to repress stem cell properties and radioresistance in non-small cell lung cancer.Apoptosis. 2023 Apr;28(3-4):397-413. doi: 10.1007/s10495-022-01789-y. Epub 2022 Nov 27. Apoptosis. 2023. PMID: 36436119
-
Six MicroRNA Prognostic Models for Overall Survival of Lung Adenocarcinoma.Genet Res (Camb). 2022 Aug 27;2022:5955052. doi: 10.1155/2022/5955052. eCollection 2022. Genet Res (Camb). 2022. PMID: 36101742 Free PMC article.
-
Extracellular vesicles and cancer stem cells: a deadly duo in tumor progression.Oncol Rev. 2024 Jul 18;18:1411736. doi: 10.3389/or.2024.1411736. eCollection 2024. Oncol Rev. 2024. PMID: 39091989 Free PMC article. Review.
-
miR-21, miR-221, and miR-222 upregulation in lung cancer promotes metastasis by reducing oxidative stress and apoptosis.Rev Assoc Med Bras (1992). 2023 Jun 2;69(6):e20221688. doi: 10.1590/1806-9282.20221688. eCollection 2023. Rev Assoc Med Bras (1992). 2023. PMID: 37283359 Free PMC article.
-
Propofol Ameliorates Microglia Activation by Targeting MicroRNA-221/222-IRF2 Axis.J Immunol Res. 2021 Aug 10;2021:3101146. doi: 10.1155/2021/3101146. eCollection 2021. J Immunol Res. 2021. PMID: 34423051 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

