Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods
- PMID: 33959631
- PMCID: PMC8093571
- DOI: 10.3389/fmolb.2021.637559
Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods
Abstract
Viruses are ubiquitous in the environment. While many impart no deleterious effects on their hosts, several are major pathogens. This risk of pathogenicity, alongside the fact that many viruses can rapidly mutate highlights the need for suitable, rapid diagnostic measures. This review provides a critical analysis of widely used methods and examines their advantages and limitations. Currently, nucleic-acid detection and immunoassay methods are among the most popular means for quickly identifying viral infection directly from source. Nucleic acid-based detection generally offers high sensitivity, but can be time-consuming, costly, and require trained staff. The use of isothermal-based amplification systems for detection could aid in the reduction of results turnaround and equipment-associated costs, making them appealing for point-of-use applications, or when high volume/fast turnaround testing is required. Alternatively, immunoassays offer robustness and reduced costs. Furthermore, some immunoassay formats, such as those using lateral-flow technology, can generate results very rapidly. However, immunoassays typically cannot achieve comparable sensitivity to nucleic acid-based detection methods. Alongside these methods, the application of next-generation sequencing can provide highly specific results. In addition, the ability to sequence large numbers of viral genomes would provide researchers with enhanced information and assist in tracing infections.
Keywords: immunoassay; isothermal amplification; next generation sequencing; nucleic-acid detection; sampling; virus.
Copyright © 2021 Cassedy, Parle-McDermott and O’Kennedy.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
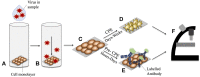
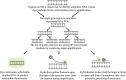


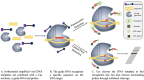
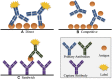
References
-
- Arunrut N., Kampeera J., Sirithammajak S., Sanguanrut P., Proespraiwong P., Suebsing R., et al. (2016). Sensitive visual detection of AHPND bacteria using loop-mediated isothermal amplification combined with DNA-Functionalized gold nanoparticles as probes. PLoS One 11, e0151769. 10.1371/journal.pone.0151769 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

