Identification of mRNA-, circRNA- and lncRNA- Associated ceRNA Networks and Potential Biomarkers for Preeclampsia From Umbilical Vein Endothelial Cells
- PMID: 33959635
- PMCID: PMC8093761
- DOI: 10.3389/fmolb.2021.652250
Identification of mRNA-, circRNA- and lncRNA- Associated ceRNA Networks and Potential Biomarkers for Preeclampsia From Umbilical Vein Endothelial Cells
Abstract
Objective: The etiology and pathogenesis of preeclampsia (PE) remain unclear, and ideal biomarkers for the early detection of PE are scarce. The involvement of the competing endogenous RNA (ceRNA) hypothesis in PE is only partially understood. The present study aimed to delineate a regulatory network in PE comprised of messenger RNAs (mRNAs), circular RNAs (circRNAs), long non-coding RNAs (lncRNAs), and microRNAs (miRNAs) via ceRNA profiles from human umbilical vein endothelial cells (HUVECs) to further reveal the pathogenesis of PE and potential biomarkers.
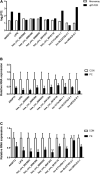
Methods: Differentially expressed mRNAs, circRNAs, and lncRNAs were detected in HUVECs from early onset preeclampsia (EOPE) cases (n = 4) and normal pregnancies (n = 4) by microarray analysis. Bioinformatics analysis was performed to systematically analyze the data, and a relevant ceRNA network was constructed. RNAs (ANGPT2, LIPG, hsa_circ_0025992, hsa_circ_0090396, hsa_circ_0066955, hsa_circ_0041203, hsa_circ_0018116, lnc-C17orf64-1:1, lnc-SLC27A2-2:1, and lnc-UEVLD-5:1) were validated by quantitative real-time PCR (qRT-PCR) in 10 pairs of HUVECs and placental tissues from PE patients and normal pregnancies. Furthermore, expression of hsa_circ_0025992 was detected in maternal peripheral blood samples from PE patients (n = 24) and normal pregnancies (n = 30) to confirm its potential as a novel biomarker. The receiver operating characteristic (ROC) curve was applied to analyze its diagnostic value.
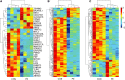
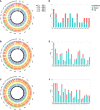
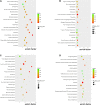
Results: Compared with HUVECs from normal pregnancies, HUVECs from EOPE cases had 33 differentially expressed mRNAs (DEmRNAs), 272 DEcircRNAs, and 207 DElncRNAs. GO and KEGG analyses of the DERNAs revealed the biological processes and pathways involved in PE. Based on the microarray data and the predicted miRNAs, a ceRNA network was constructed with four mRNAs, 34 circRNAs, nine lncRNAs, and 99 miRNAs. GO and KEGG analyses of the network reinforced the crucial roles of metabolic disorders, the p53 and JAK/STAT signaling pathways in PE. In addition, ROC analysis indicated that hsa_circ_0025992 could be used as a novel biomarker for PE.
Conclusion: A novel ceRNA network was revealed in PE, and the potential of hsa_circ_0025992 to serve as a new biomarker was confirmed.
Keywords: HUVECs; biomarker; ceRNA network; circRNAs; preeclampsia.
Copyright © 2021 Chen, He, Zheng, Wang, Zhao, Liu, Yang and Cheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- ACOG (2020). Gestational Hypertension and Preeclampsia: ACOG Practice Bulletin, Number 222. Obstet. Gynecol. 135 e237–e260. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

