Common genetic associations between age-related diseases
- PMID: 33959723
- PMCID: PMC7610725
- DOI: 10.1038/s43587-021-00051-5
Common genetic associations between age-related diseases
Abstract
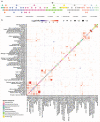
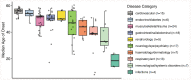
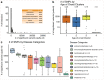
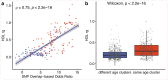
Age is a common risk factor in many diseases, but the molecular basis for this relationship is elusive. In this study we identified 4 disease clusters from 116 diseases in the UK Biobank data, defined by their age-of-onset profiles, and found that diseases with the same onset profile are genetically more similar, suggesting a common etiology. This similarity was not explained by disease categories, co-occurrences or disease cause-effect relationships. Two of the four disease clusters had an increased risk of occurrence from age 20 and 40 years respectively. They both showed an association with known aging-related genes, yet differed in functional enrichment and evolutionary profiles. Moreover, they both had age-related expression and methylation changes. We also tested mutation accumulation and antagonistic pleiotropy theories of aging and found support for both.
Keywords: Aging; GWAS; UK Biobank; age-related disease; antagonistic pleiotropy; mutation accumulation.
Conflict of interest statement
Competing interests The authors declare that they have no competing interests.
Figures






References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

