Process development and scale-up optimization of the SARS-CoV-2 receptor binding domain-based vaccine candidate, RBD219-N1C1
- PMID: 33959781
- PMCID: PMC8102132
- DOI: 10.1007/s00253-021-11281-3
Process development and scale-up optimization of the SARS-CoV-2 receptor binding domain-based vaccine candidate, RBD219-N1C1
Abstract
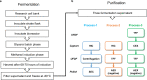
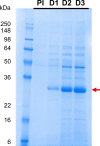
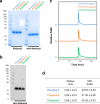
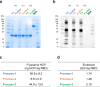
A SARS-CoV-2 RBD219-N1C1 (RBD219-N1C1) recombinant protein antigen formulated on Alhydrogel® has recently been shown to elicit a robust neutralizing antibody response against SARS-CoV-2 pseudovirus in mice. The antigen has been produced under current good manufacturing practices (cGMPs) and is now in clinical testing. Here, we report on process development and scale-up optimization for upstream fermentation and downstream purification of the antigen. This includes production at the 1-L and 5-L scales in the yeast, Pichia pastoris, and the comparison of three different chromatographic purification methods. This culminated in the selection of a process to produce RBD219-N1C1 with a yield of >400 mg per liter of fermentation with >92% purity and >39% target product recovery after purification. In addition, we show the results from analytical studies, including SEC-HPLC, DLS, and an ACE2 receptor binding assay that were performed to characterize the purified proteins to select the best purification process. Finally, we propose an optimized upstream fermentation and downstream purification process that generates quality RBD219-N1C1 protein antigen and is fully scalable at a low cost. KEY POINTS: • Yeast fermentation conditions for a recombinant COVID-19 vaccine were determined. • Three purification protocols for a COVID-19 vaccine antigen were compared. • Reproducibility of a scalable, low-cost process for a COVID-19 vaccine was shown. Graphical abstract.
Keywords: COVID-19; Fermentation; Pichia pastoris; Purification; Spike protein.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Chen WH, Chag SM, Poongavanam MV, Biter AB, Ewere EA, Rezende W, Seid CA, Hudspeth EM, Pollet J, McAtee CP, Strych U, Bottazzi ME, Hotez PJ. Optimization of the production process and characterization of the yeast-expressed SARS-CoV recombinant receptor-binding domain (RBD219-N1), a SARS vaccine candidate. J Pharm Sci. 2017;106(8):1961–1970. doi: 10.1016/j.xphs.2017.04.037. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

