Contribution of the Gut Microbiome to Drug Disposition, Pharmacokinetic and Pharmacodynamic Variability
- PMID: 33959897
- PMCID: PMC8332605
- DOI: 10.1007/s40262-021-01032-y
Contribution of the Gut Microbiome to Drug Disposition, Pharmacokinetic and Pharmacodynamic Variability
Abstract
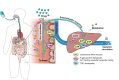
The trillions of microbes that make up the gut microbiome are an important contributor to health and disease. With respect to xenobiotics, particularly orally administered compounds, the gut microbiome interacts directly with drugs to break them down into metabolic products. In addition, microbial products such as bile acids interact with nuclear receptors on host drug-metabolizing enzyme machinery, thus indirectly influencing drug disposition and pharmacokinetics. Gut microbes also influence drugs that undergo enterohepatic recycling by reversing host enzyme metabolic processes and increasing exposure to toxic metabolites as exemplified by the chemotherapy agent irinotecan and non-steroidal anti-inflammatory drugs. Recent data with immune checkpoint inhibitors demonstrate the impact of the gut microbiome on drug pharmacodynamics. We summarize the clinical importance of gut microbe interaction with digoxin, irinotecan, immune checkpoint inhibitors, levodopa, and non-steroidal anti-inflammatory drugs. Understanding the complex interactions of the gut microbiome with xenobiotics is challenging; and highly sensitive methods such as untargeted metabolomics with molecular networking along with other in silico methods and animal and human in vivo studies will uncover mechanisms and pathways. Incorporating the contribution of the gut microbiome to drug disposition, pharmacokinetics, and pharmacodynamics is vital in this era of precision medicine.
© 2021. The Author(s).
Conflict of interest statement
Shirley M. Tsunoda, Christopher Gonzales, Alan K. Jarmusch, Jeremiah D. Momper, and Joseph D. Ma have no conflicts of interest that are directly relevant to the content of this article.
Figures
Similar articles
-
Gut Reactions: Breaking Down Xenobiotic-Microbiome Interactions.Pharmacol Rev. 2019 Apr;71(2):198-224. doi: 10.1124/pr.118.015768. Pharmacol Rev. 2019. PMID: 30890566 Review.
-
Potential Implications of Gut Microbiota in Drug Pharmacokinetics and Bioavailability.Pharmacotherapy. 2020 Jul;40(7):704-712. doi: 10.1002/phar.2428. Epub 2020 Jun 30. Pharmacotherapy. 2020. PMID: 32463481 Review.
-
Connecting the dots: Targeting the microbiome in drug toxicity.Med Res Rev. 2022 Jan;42(1):83-111. doi: 10.1002/med.21805. Epub 2021 Apr 15. Med Res Rev. 2022. PMID: 33856076 Review.
-
Gut Microbiome and Response to Cardiovascular Drugs.Circ Genom Precis Med. 2019 Sep;12(9):421-429. doi: 10.1161/CIRCGEN.119.002314. Epub 2019 Aug 28. Circ Genom Precis Med. 2019. PMID: 31462078 Free PMC article. Review.
-
Omics for Understanding the Gut-Liver-Microbiome Axis and Precision Medicine.Clin Pharmacol Drug Dev. 2017 Mar;6(2):176-185. doi: 10.1002/cpdd.310. Clin Pharmacol Drug Dev. 2017. PMID: 28263462 Review.
Cited by
-
Gut Microbiome in Colorectal Cancer: Clinical Diagnosis and Treatment.Genomics Proteomics Bioinformatics. 2023 Feb;21(1):84-96. doi: 10.1016/j.gpb.2022.07.002. Epub 2022 Jul 30. Genomics Proteomics Bioinformatics. 2023. PMID: 35914737 Free PMC article. Review.
-
Advancements in precision medicine: multi-omics approach for tailored metformin treatment in type 2 diabetes.Front Pharmacol. 2024 Nov 28;15:1506767. doi: 10.3389/fphar.2024.1506767. eCollection 2024. Front Pharmacol. 2024. PMID: 39669200 Free PMC article. Review.
-
The association between enteropathogens and antimycobacterial drug pharmacokinetics in children.Lancet Microbe. 2022 Jun;3(6):e400-e401. doi: 10.1016/S2666-5247(21)00353-0. Epub 2022 Apr 7. Lancet Microbe. 2022. PMID: 35434672 Free PMC article. No abstract available.
-
Liver-on-chips for drug discovery and development.Mater Today Bio. 2024 Jul 2;27:101143. doi: 10.1016/j.mtbio.2024.101143. eCollection 2024 Aug. Mater Today Bio. 2024. PMID: 39070097 Free PMC article. Review.
-
Oleandrin: A Systematic Review of its Natural Sources, Structural Properties, Detection Methods, Pharmacokinetics and Toxicology.Front Pharmacol. 2022 Feb 21;13:822726. doi: 10.3389/fphar.2022.822726. eCollection 2022. Front Pharmacol. 2022. PMID: 35273501 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

