A Floating Mold Technique for the Programmed Assembly of Protocells into Protocellular Materials Capable of Non-Equilibrium Biochemical Sensing
- PMID: 33960013
- PMCID: PMC11469123
- DOI: 10.1002/adma.202100340
A Floating Mold Technique for the Programmed Assembly of Protocells into Protocellular Materials Capable of Non-Equilibrium Biochemical Sensing
Abstract
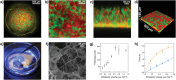
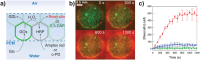
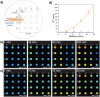
Despite important breakthroughs in bottom-up synthetic biology, a major challenge still remains the construction of free-standing, macroscopic, and robust materials from protocell building blocks that are stable in water and capable of emergent behaviors. Herein, a new floating mold technique for the fabrication of millimeter- to centimeter-sized protocellular materials (PCMs) of any shape that overcomes most of the current challenges in prototissue engineering is reported. Significantly, this technique also allows for the generation of 2D periodic arrays of PCMs that display an emergent non-equilibrium spatiotemporal sensing behavior. These arrays are capable of collectively translating the information provided by the external environment and are encoded in the form of propagating reaction-diffusion fronts into a readable dynamic signal output. Overall, the methodology opens up a route to the fabrication of macroscopic and robust tissue-like materials with emergent behaviors, providing a new paradigm of bottom-up synthetic biology and biomimetic materials science.
Keywords: bioinspiration; enzyme cascade; out-of-equilibrium; protocells; protocellular materials; prototissue.
© 2021 The Authors. Advanced Materials published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Programmed Fabrication of Vesicle-Based Prototissue Fibers with Modular Functionalities.Adv Sci (Weinh). 2025 Apr;12(16):e2409066. doi: 10.1002/advs.202409066. Epub 2025 Feb 10. Adv Sci (Weinh). 2025. PMID: 39927512 Free PMC article.
-
Bioinspired Networks of Communicating Synthetic Protocells.Front Mol Biosci. 2021 Dec 24;8:804717. doi: 10.3389/fmolb.2021.804717. eCollection 2021. Front Mol Biosci. 2021. PMID: 35004855 Free PMC article. Review.
-
Evolving protocells to prototissues: rational design of a missing link.Biochem Soc Trans. 2013 Oct;41(5):1159-65. doi: 10.1042/BST20130135. Biochem Soc Trans. 2013. PMID: 24059502 Review.
-
Programmed assembly of synthetic protocells into thermoresponsive prototissues.Nat Mater. 2018 Dec;17(12):1145-1153. doi: 10.1038/s41563-018-0183-5. Epub 2018 Oct 8. Nat Mater. 2018. PMID: 30297813
-
Matching Together Living Cells and Prototissues: Will There Be Chemistry?Chembiochem. 2024 Sep 16;25(18):e202400378. doi: 10.1002/cbic.202400378. Epub 2024 Aug 7. Chembiochem. 2024. PMID: 39031571 Review.
Cited by
-
Continuous Light-Induced Water Oxidation in Polyoxometalate-Based Photocatalytic Protocells and Prototissues.Chemistry. 2025 Jul 11;31(39):e202501322. doi: 10.1002/chem.202501322. Epub 2025 Jun 22. Chemistry. 2025. PMID: 40459204 Free PMC article.
-
High-throughput production of functional prototissues capable of producing NO for vasodilation.Nat Commun. 2022 Apr 20;13(1):2148. doi: 10.1038/s41467-022-29571-3. Nat Commun. 2022. PMID: 35444179 Free PMC article.
-
Programmed Fabrication of Vesicle-Based Prototissue Fibers with Modular Functionalities.Adv Sci (Weinh). 2025 Apr;12(16):e2409066. doi: 10.1002/advs.202409066. Epub 2025 Feb 10. Adv Sci (Weinh). 2025. PMID: 39927512 Free PMC article.
-
Dynamic satellite-parent liposome networks for quantitative microreactions.Chem Sci. 2024 Nov 6;15(46):19619-19625. doi: 10.1039/d4sc04925j. eCollection 2024 Nov 27. Chem Sci. 2024. PMID: 39568867 Free PMC article.
-
A Photo-degradable Crosslinker for the Development of Light-responsive Protocell Membranes.Chemistry. 2023 Nov 2;29(61):e202302058. doi: 10.1002/chem.202302058. Epub 2023 Sep 22. Chemistry. 2023. PMID: 37497813 Free PMC article.
References
-
- a) Liu J. S., Gartner Z. J., Trends Cell Biol. 2012, 22, 683; - PMC - PubMed
- b) Editorial, Nature 2018, 563, 155;
- c) Powell K., Nature 2018, 563, 172; - PubMed
- d) Bayley H., Cazimoglu I., Hoskin C. E. G., Life Sci. 2019, 3, 615; - PMC - PubMed
- e) Gonzales D. T., Zechner C., Tang T. Y. D., Curr. Opin. Syst. Biol. 2020, 24, 56;
- f) Gobbo P., Biochem. Soc. Trans. 2020, 48, 2579. - PubMed
-
- a) Kurihara K., Tamura M., Shohda K., Toyota T., Suzuki K., Sugawara T., Nat. Chem. 2011, 3, 775; - PubMed
- b) Dzieciol A. J., Mann S., Chem. Soc. Rev. 2012, 41, 79; - PubMed
- c) Li M., Harbron R. L., Weaver J. V., Binks B. P., Mann S., Nat. Chem. 2013, 5, 529; - PubMed
- d) Marguet M., Bonduelle C., Lecommandoux S., Chem. Soc. Rev. 2013, 42, 512; - PubMed
- e) Dora Tang T. Y., Rohaida Che Hak C., Thompson A. J., Kuimova M. K., Williams D. S., Perriman A. W., Mann S., Nat. Chem. 2014, 6, 527; - PubMed
- f) Xu C., Hu S., Chen X., Mater. Today 2016, 19, 516; - PMC - PubMed
- g) Rodriguez‐Arco L., Li M., Mann S., Nat. Mater. 2017, 16, 857. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

