Insights into the substrate discrimination mechanisms of methyl-CpG-binding domain 4
- PMID: 33960375
- PMCID: PMC8173489
- DOI: 10.1042/BCJ20210017
Insights into the substrate discrimination mechanisms of methyl-CpG-binding domain 4
Abstract
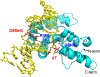
G:T mismatches, the major mispairs generated during DNA metabolism, are repaired in part by mismatch-specific DNA glycosylases such as methyl-CpG-binding domain 4 (MBD4) and thymine DNA glycosylase (TDG). Mismatch-specific DNA glycosylases must discriminate the mismatches against million-fold excess correct base pairs. MBD4 efficiently removes thymine opposite guanine but not opposite adenine. Previous studies have revealed that the substrate thymine is flipped out and enters the catalytic site of the enzyme, while the estranged guanine is stabilized by Arg468 of MBD4. To gain further insights into the mismatch discrimination mechanism of MBD4, we assessed the glycosylase activity of MBD4 toward various base pairs. In addition, we determined a crystal structure of MBD4 bound to T:O6-methylguanine-containing DNA, which suggests the O6 and N2 of purine and the O4 of pyrimidine are required to be a substrate for MBD4. To understand the role of the Arg468 finger in catalysis, we evaluated the glycosylase activity of MBD4 mutants, which revealed the guanidinium moiety of Arg468 may play an important role in catalysis. D560N/R468K MBD4 bound to T:G mismatched DNA shows that the side chain amine moiety of the Lys stabilizes the flipped-out thymine by a water-mediated phosphate pinching, while the backbone carbonyl oxygen of the Lys engages in hydrogen bonds with N2 of the estranged guanine. Comparison of various DNA glycosylase structures implies the guanidinium and amine moieties of Arg and Lys, respectively, may involve in discriminating between substrate mismatches and nonsubstrate base pairs.
Keywords: DNA glycosylase; base excision repair; substrate recognition.
© 2021 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
Figures

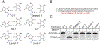

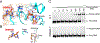



References
-
- Kunkel TA (1985) The mutational specificity of DNA polymerase-beta during in vitro DNA synthesis. Production of frameshift, base substitution, and deletion mutations. J Biol Chem 260, 5787–5796 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

