Protein-DNA/RNA Interactions: An Overview of Investigation Methods in the -Omics Era
- PMID: 33961438
- PMCID: PMC8280749
- DOI: 10.1021/acs.jproteome.1c00074
Protein-DNA/RNA Interactions: An Overview of Investigation Methods in the -Omics Era
Abstract
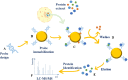
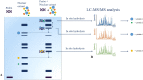
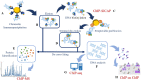
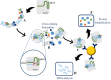
The fields of application of functional proteomics are not limited to the study of protein-protein interactions; they also extend to those involving protein complexes that bind DNA or RNA. These interactions affect fundamental processes such as replication, transcription, and repair in the case of DNA, as well as transport, translation, splicing, and silencing in the case of RNA. Analytical or preparative experimental approaches, both in vivo and in vitro, have been developed to isolate and identify DNA/RNA binding proteins by exploiting the advantage of the affinity shown by these proteins toward a specific oligonucleotide sequence. The present review proposes an overview of the approaches most commonly employed in proteomics applications for the identification of nucleic acid-binding proteins, such as affinity purification (AP) protocols, EMSA, chromatin purification methods, and CRISPR-based chromatin affinity purification, which are generally associated with mass spectrometry methodologies for the unbiased protein identification.
Keywords: CRISPR-Cas9; ChIP; DNA−protein interactions; EMSA; RNA−protein interactions; mass spectrometry; proteomics; pull-down.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- Zuckerkandl E.; Pauling L. B.. Molecular disease, evolution, and genetic heterogeneity. In Horizons in Biochemistry; Kasha M., Pullman B., Eds.; Academic Press: New York, 1962; pp 189–225.
-
- Preeti P.; Sabeeha H.; Shandar A., Protein-DNA Interactions. In Encyclopedia of Bioinformatics and Computational Biology; Academic Press, 2019; pp 142–154.
-
- Chowdhury N.; Bagchi A. An Overview of DNA-Protein Interactions. Curr. Chem. Biol. 2016, 9, 73.10.2174/2212796809666151022202255. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

