Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2
- PMID: 33961632
- PMCID: PMC8133773
- DOI: 10.1371/journal.pbio.3001236
Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2
Abstract
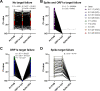
With the emergence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) variants that may increase transmissibility and/or cause escape from immune responses, there is an urgent need for the targeted surveillance of circulating lineages. It was found that the B.1.1.7 (also 501Y.V1) variant, first detected in the United Kingdom, could be serendipitously detected by the Thermo Fisher TaqPath COVID-19 PCR assay because a key deletion in these viruses, spike Δ69-70, would cause a "spike gene target failure" (SGTF) result. However, a SGTF result is not definitive for B.1.1.7, and this assay cannot detect other variants of concern (VOC) that lack spike Δ69-70, such as B.1.351 (also 501Y.V2), detected in South Africa, and P.1 (also 501Y.V3), recently detected in Brazil. We identified a deletion in the ORF1a gene (ORF1a Δ3675-3677) in all 3 variants, which has not yet been widely detected in other SARS-CoV-2 lineages. Using ORF1a Δ3675-3677 as the primary target and spike Δ69-70 to differentiate, we designed and validated an open-source PCR assay to detect SARS-CoV-2 VOC. Our assay can be rapidly deployed in laboratories around the world to enhance surveillance for the local emergence and spread of B.1.1.7, B.1.351, and P.1.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Update of
-
PCR assay to enhance global surveillance for SARS-CoV-2 variants of concern.medRxiv [Preprint]. 2021 Mar 12:2021.01.28.21250486. doi: 10.1101/2021.01.28.21250486. medRxiv. 2021. Update in: PLoS Biol. 2021 May 7;19(5):e3001236. doi: 10.1371/journal.pbio.3001236. PMID: 33758901 Free PMC article. Updated. Preprint.
References
-
- Davies NG, Barnard RC, Jarvis CI, Kucharski AJ, Munday J, Pearson CAB, et al. Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. medRxiv. 2020. 10.1101/2020.12.24.20248822 - DOI
-
- Rambaut A, Loman N, Pybus O BW, Barrett J, Carabelli A, Connor T, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. 2020. December [cited 2021 Jan 23]. Available from: https://virological.org/t/preliminary-genomic-characterisation-of-an-eme...
-
- Borges V, Sousa C, Menezes L, A MG, Picão M, Almeida JP, et al. Tracking SARS-CoV-2 VOC 202012/01 (lineage B.1.1.7) dissemination in Portugal: insights from nationwide RT-PCR Spike gene drop out data. 2021. January [cited 2021 Jan 23]. Available from: https://virological.org/t/tracking-sars-cov-2-voc-202012-01-lineage-b-1-... - PMC - PubMed
-
- Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv. 2021. 10.1101/2020.12.30.20249034 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

