Microbial co-infections in COVID-19: Associated microbiota and underlying mechanisms of pathogenesis
- PMID: 33962007
- PMCID: PMC8095020
- DOI: 10.1016/j.micpath.2021.104941
Microbial co-infections in COVID-19: Associated microbiota and underlying mechanisms of pathogenesis
Abstract
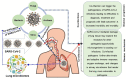
The novel coronavirus infectious disease-2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has traumatized the whole world with the ongoing devastating pandemic. A plethora of microbial domains including viruses (other than SARS-CoV-2), bacteria, archaea and fungi have evolved together, and interact in complex molecular pathogenesis along with SARS-CoV-2. However, the involvement of other microbial co-pathogens and underlying molecular mechanisms leading to extortionate ailment in critically ill COVID-19 patients has yet not been extensively reviewed. Although, the incidence of co-infections could be up to 94.2% in laboratory-confirmed COVID-19 cases, the fate of co-infections among SARS-CoV-2 infected hosts often depends on the balance between the host's protective immunity and immunopathology. Predominantly identified co-pathogens of SARS-CoV-2 are bacteria such as Streptococcus pneumoniae, Staphylococcus aureus, Klebsiella pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae, Acinetobacter baumannii, Legionella pneumophila and Clamydia pneumoniae followed by viruses including influenza, coronavirus, rhinovirus/enterovirus, parainfluenza, metapneumovirus, influenza B virus, and human immunodeficiency virus. The cross-talk between co-pathogens (especially lung microbiomes), SARS-CoV-2 and host is an important factor that ultimately increases the difficulty of diagnosis, treatment, and prognosis of COVID-19. Simultaneously, co-infecting microbiotas may use new strategies to escape host defense mechanisms by altering both innate and adaptive immune responses to further aggravate SARS-CoV-2 pathogenesis. Better understanding of co-infections in COVID-19 is critical for the effective patient management, treatment and containment of SARS-CoV-2. This review therefore necessitates the comprehensive investigation of commonly reported microbial co-pathogens amid COVID-19, their transmission pattern along with the possible mechanism of co-infections and outcomes. Thus, identifying the possible co-pathogens and their underlying molecular mechanisms during SARS-CoV-2 pathogenesis may shed light in developing diagnostics, appropriate curative and preventive interventions for suspected SARS-CoV-2 respiratory infections in the current pandemic.
Keywords: COVID-19; Microbial co-infections; Molecular pathogenesis; SARS-CoV-2.
Copyright © 2021. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare no conflict of interests.
Figures
Similar articles
-
Co-infections among patients with COVID-19: The need for combination therapy with non-anti-SARS-CoV-2 agents?J Microbiol Immunol Infect. 2020 Aug;53(4):505-512. doi: 10.1016/j.jmii.2020.05.013. Epub 2020 May 23. J Microbiol Immunol Infect. 2020. PMID: 32482366 Free PMC article. Review.
-
Coinfections with Other Respiratory Pathogens among Patients with COVID-19.Microbiol Spectr. 2021 Sep 3;9(1):e0016321. doi: 10.1128/Spectrum.00163-21. Epub 2021 Jul 21. Microbiol Spectr. 2021. PMID: 34287033 Free PMC article.
-
Decoding the double trouble: A mathematical modelling of co-infection dynamics of SARS-CoV-2 and influenza-like illness.Biosystems. 2023 Feb;224:104827. doi: 10.1016/j.biosystems.2023.104827. Epub 2023 Jan 7. Biosystems. 2023. PMID: 36626949 Free PMC article.
-
Bacterial co-infections with SARS-CoV-2.IUBMB Life. 2020 Oct;72(10):2097-2111. doi: 10.1002/iub.2356. Epub 2020 Aug 8. IUBMB Life. 2020. PMID: 32770825 Free PMC article. Review.
-
Analysis of coexisting pathogens in nasopharyngeal swabs from COVID-19.Front Cell Infect Microbiol. 2023 Jun 22;13:1140548. doi: 10.3389/fcimb.2023.1140548. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37424777 Free PMC article.
Cited by
-
Mathematical Modeling of the Lethal Synergism of Coinfecting Pathogens in Respiratory Viral Infections: A Review.Microorganisms. 2023 Dec 13;11(12):2974. doi: 10.3390/microorganisms11122974. Microorganisms. 2023. PMID: 38138118 Free PMC article. Review.
-
SARS-CoV-2-Legionella Co-Infections: A Systematic Review and Meta-Analysis (2020-2021).Microorganisms. 2022 Feb 23;10(3):499. doi: 10.3390/microorganisms10030499. Microorganisms. 2022. PMID: 35336074 Free PMC article. Review.
-
From Co-Infections to Autoimmune Disease via Hyperactivated Innate Immunity: COVID-19 Autoimmune Coagulopathies, Autoimmune Myocarditis and Multisystem Inflammatory Syndrome in Children.Int J Mol Sci. 2023 Feb 3;24(3):3001. doi: 10.3390/ijms24033001. Int J Mol Sci. 2023. PMID: 36769320 Free PMC article. Review.
-
Therapeutic and palliative role of a Unani herbal decoction in COVID-19 and similar respiratory viral illnesses: Phytochemical & pharmacological perspective.J Ethnopharmacol. 2022 Oct 28;297:115526. doi: 10.1016/j.jep.2022.115526. Epub 2022 Jul 14. J Ethnopharmacol. 2022. PMID: 35843408 Free PMC article. Review.
-
SARS-CoV-2 Related Viral Respiratory Co-Infections: A Narrative Review.Tanaffos. 2023 Jan;22(1):19-26. Tanaffos. 2023. PMID: 37920316 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

