A survey of the mouse hindbrain in the fed and fasted states using single-nucleus RNA sequencing
- PMID: 33962048
- PMCID: PMC8170503
- DOI: 10.1016/j.molmet.2021.101240
A survey of the mouse hindbrain in the fed and fasted states using single-nucleus RNA sequencing
Abstract
Objective: The area postrema (AP) and nucleus tractus solitarius (NTS) located in the hindbrain are key nuclei that sense and integrate peripheral nutritional signals and consequently regulate feeding behaviour. While single-cell transcriptomics have been used in mice to reveal the gene expression profile and heterogeneity of key hypothalamic populations, similar in-depth studies have not yet been performed in the hindbrain.
Methods: Using single-nucleus RNA sequencing, we provide a detailed survey of 16,034 cells within the AP and NTS of mice in the fed and fasted states.
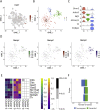
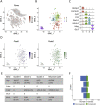
Results: Of these, 8,910 were neurons that group into 30 clusters, with 4,289 from mice fed ad libitum and 4,621 from overnight fasted mice. A total of 7,124 nuclei were from non-neuronal cells, including oligodendrocytes, astrocytes, and microglia. Interestingly, we identified that the oligodendrocyte population was particularly transcriptionally sensitive to an overnight fast. The receptors GLP1R, GIPR, GFRAL, and CALCR, which bind GLP1, GIP, GDF15, and amylin, respectively, are all expressed in the hindbrain and are major targets for anti-obesity therapeutics. We characterise the transcriptomes of these four populations and show that their gene expression profiles are not dramatically altered by an overnight fast. Notably, we find that roughly half of cells that express GIPR are oligodendrocytes. Additionally, we profile POMC-expressing neurons within the hindbrain and demonstrate that 84% of POMC neurons express either PCSK1, PSCK2, or both, implying that melanocortin peptides are likely produced by these neurons.
Conclusion: We provide a detailed single-cell level characterisation of AP and NTS cells expressing receptors for key anti-obesity drugs that are either already approved for human use or in clinical trials. This resource will help delineate the mechanisms underlying the effectiveness of these compounds and also prove useful in the continued search for other novel therapeutic targets.
Keywords: Area postrema; Gene expression; Nucleus tractus solitarius; Obesity; Therapeutics.
Copyright © 2021 The Author(s). Published by Elsevier GmbH.. All rights reserved.
Figures





References
-
- Price C.J., Hoyda T.D., Ferguson A.V. The area postrema: a brain monitor and integrator of systemic autonomic state. The Neuroscientist. 2007;14(2):182–194. - PubMed
-
- Pi-Sunyer X., Astrup A., Fujioka K., Greenway F., Halpern A., Krempf M. A randomized, controlled trial of 3.0 mg of liraglutide in weight management. New England Journal of Medicine. 2015;373(1):11–22. - PubMed
-
- Wilding J.P.H., Batterham R.L., Calanna S., Davies M., Van Gaal L.F., Lingvay I. Once-weekly semaglutide in adults with overweight or obesity. New England Journal of Medicine. 2021 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

