A diverse member of the fungal Avr4 effector family interacts with de-esterified pectin in plant cell walls to disrupt their integrity
- PMID: 33962956
- PMCID: PMC8104879
- DOI: 10.1126/sciadv.abe0809
A diverse member of the fungal Avr4 effector family interacts with de-esterified pectin in plant cell walls to disrupt their integrity
Abstract
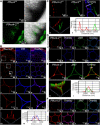
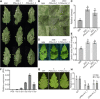
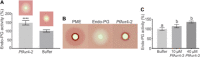
Effectors are small, secreted proteins that promote pathogen virulence. Although key to microbial infections, unlocking the intrinsic function of effectors remains a challenge. We have previously shown that members of the fungal Avr4 effector family use a carbohydrate-binding module of family 14 (CBM14) to bind chitin in fungal cell walls and protect them from host chitinases during infection. Here, we show that gene duplication in the Avr4 family produced an Avr4-2 paralog with a previously unknown effector function. Specifically, we functionally characterize PfAvr4-2, a paralog of PfAvr4 in the tomato pathogen Pseudocercospora fuligena, and show that although it contains a CBM14 domain, it does not bind chitin or protect fungi against chitinases. Instead, PfAvr4-2 interacts with highly de-esterified pectin in the plant's middle lamellae or primary cell walls and interferes with Ca2+-mediated cross-linking at cell-cell junction zones, thus loosening the plant cell wall structure and synergizing the activity of pathogen secreted endo-polygalacturonases.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures





References
-
- Jones J. D. G., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). - PubMed
-
- Bacete L., Mélida H., Miedes E., Molina A., Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 93, 614–636 (2018). - PubMed
-
- De Lorenzo G., Ferrari S., Giovannoni M., Mattei B., Cervone F., Cell wall traits that influence plant development, immunity, and bioconversion. Plant J. 97, 134–147 (2019). - PubMed
-
- Caffall K. H., Mohnen D., The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344, 1879–1900 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

