Subtelomeric assembly of a multi-gene pathway for antimicrobial defense compounds in cereals
- PMID: 33963185
- PMCID: PMC8105312
- DOI: 10.1038/s41467-021-22920-8
Subtelomeric assembly of a multi-gene pathway for antimicrobial defense compounds in cereals
Abstract
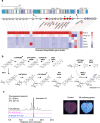
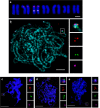
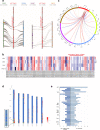
Non-random gene organization in eukaryotes plays a significant role in genome evolution. Here, we investigate the origin of a biosynthetic gene cluster for production of defence compounds in oat-the avenacin cluster. We elucidate the structure and organisation of this 12-gene cluster, characterise the last two missing pathway steps, and reconstitute the entire pathway in tobacco by transient expression. We show that the cluster has formed de novo since the divergence of oats in a subtelomeric region of the genome that lacks homology with other grasses, and that gene order is approximately colinear with the biosynthetic pathway. We speculate that the positioning of the late pathway genes furthest away from the telomere may mitigate against a 'self-poisoning' scenario in which toxic intermediates accumulate as a result of telomeric gene deletions. Our investigations reveal a striking example of adaptive evolution underpinned by remarkable genome plasticity.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Turner EM. The nature of resistance of oats to the take-all fungus. III. Distribution of the inhibitor in oat seedlings. J. Exp. Bot. 1960;11:403–412. doi: 10.1093/jxb/11.3.403. - DOI
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/J/00000614/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/K005952/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L014130/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/000PR9790/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- U01 GM110699/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

