Marker-free quantification of repair pathway utilization at Cas9-induced double-strand breaks
- PMID: 33963863
- PMCID: PMC8136827
- DOI: 10.1093/nar/gkab299
Marker-free quantification of repair pathway utilization at Cas9-induced double-strand breaks
Abstract
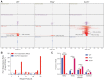
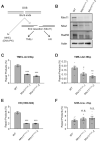
Genome integrity and genome engineering require efficient repair of DNA double-strand breaks (DSBs) by non-homologous end joining (NHEJ), homologous recombination (HR), or alternative end-joining pathways. Here we describe two complementary methods for marker-free quantification of DSB repair pathway utilization at Cas9-targeted chromosomal DSBs in mammalian cells. The first assay features the analysis of amplicon next-generation sequencing data using ScarMapper, an iterative break-associated alignment algorithm to classify individual repair products based on deletion size, microhomology usage, and insertions. The second assay uses repair pathway-specific droplet digital PCR assays ('PathSig-dPCR') for absolute quantification of signature DSB repair outcomes. We show that ScarMapper and PathSig-dPCR enable comprehensive assessment of repair pathway utilization in different cell models, after a variety of experimental perturbations. We use these assays to measure the differential impact of DNA end resection on NHEJ, HR and polymerase theta-mediated end joining (TMEJ) repair. These approaches are adaptable to any cellular model system and genomic locus where Cas9-mediated targeting is feasible. Thus, ScarMapper and PathSig-dPCR allow for systematic fate mapping of a targeted DSB with facile and accurate quantification of DSB repair pathway choice at endogenous chromosomal loci.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



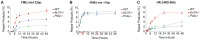
Similar articles
-
Measuring nonhomologous end-joining, homologous recombination and alternative end-joining simultaneously at an endogenous locus in any transfectable human cell.Nucleic Acids Res. 2021 Jul 21;49(13):e74. doi: 10.1093/nar/gkab262. Nucleic Acids Res. 2021. PMID: 33877327 Free PMC article.
-
Analysis of chromatid-break-repair detects a homologous recombination to non-homologous end-joining switch with increasing load of DNA double-strand breaks.Mutat Res Genet Toxicol Environ Mutagen. 2021 Jul;867:503372. doi: 10.1016/j.mrgentox.2021.503372. Epub 2021 Jun 12. Mutat Res Genet Toxicol Environ Mutagen. 2021. PMID: 34266628
-
Development of a novel method to create double-strand break repair fingerprints using next-generation sequencing.DNA Repair (Amst). 2015 Feb;26:44-53. doi: 10.1016/j.dnarep.2014.12.002. Epub 2014 Dec 19. DNA Repair (Amst). 2015. PMID: 25547252
-
Microhomology-mediated end joining: Good, bad and ugly.Mutat Res. 2018 May;809:81-87. doi: 10.1016/j.mrfmmm.2017.07.002. Epub 2017 Jul 16. Mutat Res. 2018. PMID: 28754468 Free PMC article. Review.
-
DNA polymerase θ (POLQ), double-strand break repair, and cancer.DNA Repair (Amst). 2016 Aug;44:22-32. doi: 10.1016/j.dnarep.2016.05.003. Epub 2016 May 14. DNA Repair (Amst). 2016. PMID: 27264557 Free PMC article. Review.
Cited by
-
Distinct functions of PAXX and MRI during chromosomal end joining.bioRxiv [Preprint]. 2024 Aug 22:2024.08.21.607864. doi: 10.1101/2024.08.21.607864. bioRxiv. 2024. Update in: iScience. 2025 May 22;28(6):112722. doi: 10.1016/j.isci.2025.112722. PMID: 39229097 Free PMC article. Updated. Preprint.
-
Regulation of gene editing using T-DNA concatenation.Nat Plants. 2023 Sep;9(9):1398-1408. doi: 10.1038/s41477-023-01495-w. Epub 2023 Aug 31. Nat Plants. 2023. PMID: 37653336 Free PMC article.
-
Poly(ADP) ribose polymerase promotes DNA polymerase theta-mediated end joining by activation of end resection.Nat Commun. 2022 Aug 4;13(1):4547. doi: 10.1038/s41467-022-32166-7. Nat Commun. 2022. PMID: 35927262 Free PMC article.
-
Stepwise requirements for polymerases δ and θ in theta-mediated end joining.Nature. 2023 Nov;623(7988):836-841. doi: 10.1038/s41586-023-06729-7. Epub 2023 Nov 15. Nature. 2023. PMID: 37968395 Free PMC article.
-
Refined DNA repair manipulation enables a universal knock-in strategy in mouse embryos.Nat Commun. 2025 Jul 15;16(1):6502. doi: 10.1038/s41467-025-61696-z. Nat Commun. 2025. PMID: 40664653 Free PMC article.
References
-
- Yeh C.D., Richardson C.D., Corn J.E.. Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 2019; 21:1468–1478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

