Reactome and the Gene Ontology: digital convergence of data resources
- PMID: 33964129
- PMCID: PMC8504636
- DOI: 10.1093/bioinformatics/btab325
Reactome and the Gene Ontology: digital convergence of data resources
Abstract
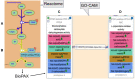
Motivation: Gene Ontology Causal Activity Models (GO-CAMs) assemble individual associations of gene products with cellular components, molecular functions and biological processes into causally linked activity flow models. Pathway databases such as the Reactome Knowledgebase create detailed molecular process descriptions of reactions and assemble them, based on sharing of entities between individual reactions into pathway descriptions.
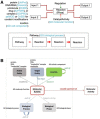
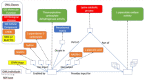
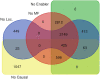
Results: To convert the rich content of Reactome into GO-CAMs, we have developed a software tool, Pathways2GO, to convert the entire set of normal human Reactome pathways into GO-CAMs. This conversion yields standard GO annotations from Reactome content and supports enhanced quality control for both Reactome and GO, yielding a nearly seamless conversion between these two resources for the bioinformatics community.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2021. Published by Oxford University Press.
Figures
References
-
- Balhoff J.P. et al. (2018) Arachne: an OWL RL reasoner applied to Gene Ontology Causal Activity Models (and beyond). In International Semantic Web Conference (P&D/Industry/BlueSky).
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

