Parental genome unification is highly error-prone in mammalian embryos
- PMID: 33964210
- PMCID: PMC8162515
- DOI: 10.1016/j.cell.2021.04.013
Parental genome unification is highly error-prone in mammalian embryos
Abstract
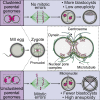
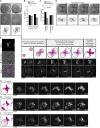
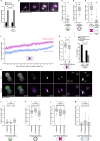
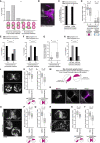
Most human embryos are aneuploid. Aneuploidy frequently arises during the early mitotic divisions of the embryo, but its origin remains elusive. Human zygotes that cluster their nucleoli at the pronuclear interface are thought to be more likely to develop into healthy euploid embryos. Here, we show that the parental genomes cluster with nucleoli in each pronucleus within human and bovine zygotes, and clustering is required for the reliable unification of the parental genomes after fertilization. During migration of intact pronuclei, the parental genomes polarize toward each other in a process driven by centrosomes, dynein, microtubules, and nuclear pore complexes. The maternal and paternal chromosomes eventually cluster at the pronuclear interface, in direct proximity to each other, yet separated. Parental genome clustering ensures the rapid unification of the parental genomes on nuclear envelope breakdown. However, clustering often fails, leading to chromosome segregation errors and micronuclei, incompatible with healthy embryo development.
Keywords: aneuploidy; centrosomes; chromosome segregation; dynein; fertilization; genome organization; human embryo; microtubules; nuclear pore complex; spindle.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures










References
-
- Arlucea J., Andrade R., Alonso R., Aréchaga J. The nuclear basket of the nuclear pore complex is part of a higher-order filamentous network that is related to chromatin. J. Struct. Biol. 1998;124:51–58. - PubMed
-
- Beaudouin J., Gerlich D., Daigle N., Eils R., Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. - PubMed
-
- Benkhalifa M., Kasakyan S., Clement P., Baldi M., Tachdjian G., Demirol A., Gurgan T., Fiorentino F., Mohammed M., Qumsiyeh M.B. Array comparative genomic hybridization profiling of first-trimester spontaneous abortions that fail to grow in vitro. Prenat. Diagn. 2005;25:894–900. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

