Monitoring of diverse enteric pathogens across environmental and host reservoirs with TaqMan array cards and standard qPCR: a methodological comparison study
- PMID: 33964239
- PMCID: PMC8116308
- DOI: 10.1016/S2542-5196(21)00051-6
Monitoring of diverse enteric pathogens across environmental and host reservoirs with TaqMan array cards and standard qPCR: a methodological comparison study
Erratum in
-
Correction Lancet Planet Health 2021; 5: e297-308.Lancet Planet Health. 2021 Jun;5(6):e336. doi: 10.1016/S2542-5196(21)00139-X. Epub 2021 May 19. Lancet Planet Health. 2021. PMID: 34019800 Free PMC article. No abstract available.
Abstract
Background: Multiple bacteria, viruses, protists, and helminths cause enteric infections that greatly impact human health and wellbeing. These enteropathogens are transmited via several pathways through human, animal, and environmental reservoirs. Individual qPCR assays have been extensively used to detect enteropathogens within these types of samples, whereas the TaqMan array card (TAC), which allows simultaneous detection of multiple enteropathogens, has only previously been validated in human clinical samples.
Methods: In this methodological comparison study, we compared the performance of a custom 48-singleplex TAC relative to standard qPCR. We established the sensitivity and specificity of each method for the detection of eight enteric targets, by using spiked samples with varying levels of PCR inhibition. We then tested the prevalence and abundance of pathogens in wastewater from Melbourne (Australia), and human, animal, and environmental samples from informal settlements in Suva, Fiji using both TAC and qPCR.
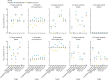
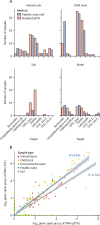
Findings: Both methods exhibited similarly h specificity (TAC 100%, qPCR 94%), sensitivity (TAC 92%, qPCR 100%), and quantitation accuracy (TAC 91%, qPCR 99%) in non-inhibited sample matrices with spiked gene fragments. PCR inhibitors substantially affected detection via TAC, though this issue was alleviated by ten-fold sample dilution. Among samples from informal settlements, the two techniques performed similarly for detection (89% agreement) and quantitation (R2 0·82) for the eight enteropathogen targets. The TAC additionally included 38 other enteric targets, enabling detection of diverse faecal pathogens and extensive environmental contamination that would be prohibitively labour intensive to assay by standard qPCR.
Interpretation: The two techniques produced similar results across diverse sample types, with qPCR prioritising greater sensitivity and quantitation accuracy, and TAC trading small reductions in these for a cost-effective larger enteropathogen panel enabling a greater number of enteric pathogens to be analysed concurrently, which is beneficial given the abundance and variety of enteric pathogens in environments such as urban informal settlements. The ability to monitor multiple enteric pathogens across diverse reservoirs could allow better resolution of pathogen exposure pathways, and the design and monitoring of interventions to reduce pathogen load.
Funding: Wellcome Trust Our Planet, Our Health programme.
Copyright © 2021 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY NC-ND 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests We declare no competing interests.
Figures

References
-
- Nadimpalli ML, Marks SJ, Montealegre MC. Urban informal settlements as hotspots of antimicrobial resistance and the need to curb environmental transmission. Nat Microbiol. 2020;5:787–795. - PubMed
-
- Liu L, Oza S, Hogan D. Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet. 2015;385:430–440. - PubMed
-
- Kotloff KL, Nataro JP, Blackwelder WC. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

