A Validated Functional Analysis of Partner and Localizer of BRCA2 Missense Variants for Use in Clinical Variant Interpretation
- PMID: 33964450
- PMCID: PMC8491091
- DOI: 10.1016/j.jmoldx.2021.04.010
A Validated Functional Analysis of Partner and Localizer of BRCA2 Missense Variants for Use in Clinical Variant Interpretation
Abstract
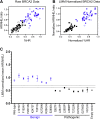
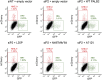
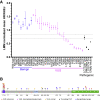
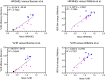
Clinical genetic testing readily detects germline genetic variants. Yet, the rarity of individual variants limits the evidence available for variant classification, leading to many variants of uncertain significance (VUS). VUS cannot guide clinical decisions, complicating counseling and management. In hereditary breast cancer gene PALB2, approximately 50% of clinically identified germline variants are VUS and approximately 90% of VUS are missense. Truncating PALB2 variants have homologous recombination (HR) defects and rely on error-prone nonhomologous end-joining for DNA damage repair (DDR). Recent reports show that some missense PALB2 variants may also be damaging, but most functional studies have lacked benchmarking controls required for sufficient predictive power for clinical use. Here, variant-level DDR capacity in hereditary breast cancer genes was assessed using the Traffic Light Reporter (TLR) to quantify cellular HR/nonhomologous end-joining with fluorescent markers. First, using BRCA2 missense variants of known significance as benchmarks, the TLR distinguished between normal/abnormal HR function. The TLR was then validated for PALB2 and used to test 37 PALB2 variants. Based on the TLR's ability to correctly classify PALB2 validation controls, these functional data where applied in subsequent germline variant interpretations at a moderate level of evidence toward a pathogenic interpretation (PS3_moderate) for 8 variants with abnormal DDR, or a supporting level of evidence toward a benign interpretation (BS3_supporting) for 13 variants with normal DDR.
Copyright © 2021 Association for Molecular Pathology and American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Functional characterization of 84 PALB2 variants of uncertain significance.Genet Med. 2020 Mar;22(3):622-632. doi: 10.1038/s41436-019-0682-z. Epub 2019 Oct 21. Genet Med. 2020. PMID: 31636395 Free PMC article.
-
Missense PALB2 germline variant disrupts nuclear localization of PALB2 in a patient with breast cancer.Fam Cancer. 2020 Apr;19(2):123-131. doi: 10.1007/s10689-020-00163-8. Fam Cancer. 2020. PMID: 32048105
-
Pathogenic and likely pathogenic variants in PALB2, CHEK2, and other known breast cancer susceptibility genes among 1054 BRCA-negative Hispanics with breast cancer.Cancer. 2019 Aug 15;125(16):2829-2836. doi: 10.1002/cncr.32083. Epub 2019 Jun 17. Cancer. 2019. PMID: 31206626 Free PMC article.
-
PALB2 Variants: Protein Domains and Cancer Susceptibility.Trends Cancer. 2021 Mar;7(3):188-197. doi: 10.1016/j.trecan.2020.10.002. Epub 2020 Nov 1. Trends Cancer. 2021. PMID: 33139182 Review.
-
The Role of PALB2 in the DNA Damage Response and Cancer Predisposition.Int J Mol Sci. 2017 Aug 31;18(9):1886. doi: 10.3390/ijms18091886. Int J Mol Sci. 2017. PMID: 28858227 Free PMC article. Review.
Cited by
-
Splicing predictions, minigene analyses, and ACMG-AMP clinical classification of 42 germline PALB2 splice-site variants.J Pathol. 2022 Mar;256(3):321-334. doi: 10.1002/path.5839. Epub 2021 Dec 28. J Pathol. 2022. PMID: 34846068 Free PMC article.
References
-
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H.L. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. - PMC - PubMed
-
- Evans J.P., Powell B.C., Berg J.S. Finding the rare pathogenic variants in a human genome. JAMA. 2017;317:1904–1905. - PubMed
-
- Guidugli L., Pankratz V.S., Singh N., Thompson J., Erding C.A., Engel C., Schmutzler R., Domchek S., Nathanson K., Radice P., Singer C., Tonin P.N., Lindor N.M., Goldgar D.E., Couch F.J. A classification model for BRCA2 DNA binding domain missense variants based on homology-directed repair activity. Cancer Res. 2013;73:265–275. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

