CXCR4 intracellular protein promotes drug resistance and tumorigenic potential by inversely regulating the expression of Death Receptor 5
- PMID: 33966046
- PMCID: PMC8106681
- DOI: 10.1038/s41419-021-03730-8
CXCR4 intracellular protein promotes drug resistance and tumorigenic potential by inversely regulating the expression of Death Receptor 5
Abstract
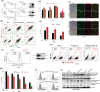
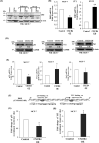
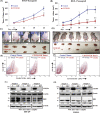
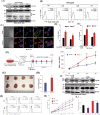
Chemokine receptor CXCR4 overexpression in solid tumors has been strongly associated with poor prognosis and adverse clinical outcome. However, blockade of CXCL12-CXCR4 signaling axis by inhibitors like Nox-A12, FDA approved CXCR4 inhibitor drug AMD3100 have shown limited clinical success in cancer treatment. Therefore, exclusive contribution of CXCR4-CXCL12 signaling in pro-tumorigenic function is questionable. In our pursuit to understand the impact of chemokine signaling in carcinogenesis, we reveal that instead of CXCR4-CXCL12 signaling, presence of CXCR4 intracellular protein augments paclitaxel resistance and pro-tumorigenic functions. In search of pro-apoptotic mechanisms for CXCR4 mediated drug resistance; we discover that DR5 is a new selective target of CXCR4 in breast and colon cancer. Further, we detect that CXCR4 directs the differential recruitment of transcription factors p53 and YY1 to the promoter of DR5 in course of its transcriptional repression. Remarkably, inhibiting CXCR4-ligand-mediated signals completely fails to block the above phenotype. Overexpression of different mutant versions of CXCR4 lacking signal transduction capabilities also result in marked downregulation of DR5 expression in colon cancer indeed confirms the reverse relationship between DR5 and intracellular CXCR4 protein expression. Irrespective of CXCR4 surface expression, by utilizing stable gain and loss of function approaches, we observe that intracellular CXCR4 protein selectively resists and sensitizes colon cancer cells against paclitaxel therapy in vitro and in vivo. Finally, performing TCGA data mining and using human breast cancer patient samples, we demonstrate that expression of CXCR4 and DR5 are inversely regulated. Together, our data suggest that targeting CXCR4 intracellular protein may be critical to dampen the pro-tumorigenic functions of CXCR4.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer J. clinicians. 2018;68:394–424. - PubMed
-
- Hanahan D., Weinberg R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). - PubMed
-
- Hata AN, Engelman JA, Faber AC. The BCL2 family: key mediators of the apoptotic response to targeted anticancer therapeutics. Cancer Disco. 2015;5:475–487. doi: 10.1158/2159-8290.CD-15-0011. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

