Making Choices: DNA Replication Fork Recovery Mechanisms
- PMID: 33967572
- PMCID: PMC8098667
- DOI: 10.1016/j.semcdb.2020.10.001
Making Choices: DNA Replication Fork Recovery Mechanisms
Abstract
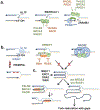
DNA replication is laden with obstacles that slow, stall, collapse, and break DNA replication forks. At each obstacle, there is a decision to be made whether to bypass the lesion, repair or restart the damaged fork, or to protect stalled forks from further demise. Each "decision" draws upon multitude of proteins participating in various mechanisms that allow repair and restart of replication forks. Specific functions for many of these proteins have been described and an understanding of how they come together in supporting replication forks is starting to emerge. Many questions, however, remain regarding selection of the mechanisms that enable faithful genome duplication and how "normal" intermediates in these mechanisms are sometimes funneled into "rogue" processes that destabilize the genome and lead to cancer, cell death, and emergence of chemotherapeutic resistance. In this review we will discuss molecular mechanisms of DNA damage bypass and replication fork protection and repair. We will specifically focus on the key players that define which mechanism is employed including: PCNA and its control by posttranslational modifications, translesion synthesis DNA polymerases, molecular motors that catalyze reversal of stalled replication forks, proteins that antagonize fork reversal and protect reversed forks from nucleolytic degradation, and the machinery of homologous recombination that helps to reestablish broken forks. We will also discuss risks to genome integrity inherent in each of these mechanisms.
Keywords: BRCA2; DNA replication; HLTF; PCNA; RAD51; RAD52; RPA; SHPRH; SMARCAL1; ZRANB3; genome stability; replication fork protection; replication fork reversal; template switching; translesion synthesis; translesion synthesis DNA polymerases.
Figures
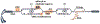
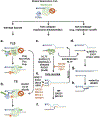
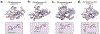
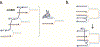
References
-
- Berti M, Cortez D, and Lopes M (2020) The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nature reviews. Molecular cell biology - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
