Characterization of a High Hierarchical Regulator, PtrGATA12, Functioning in Differentially Regulating Secondary Wall Component Biosynthesis in Populus trichocarpa
- PMID: 33968111
- PMCID: PMC8096934
- DOI: 10.3389/fpls.2021.657787
Characterization of a High Hierarchical Regulator, PtrGATA12, Functioning in Differentially Regulating Secondary Wall Component Biosynthesis in Populus trichocarpa
Abstract
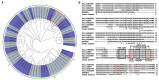
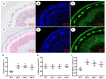
In plants, GATA transcription factors (TFs) have been reported to play vital roles in to a wide range of biological processes. To date, there is still no report about the involvement and functions of woody plant GATA TFs in wood formation. In this study, we described the functional characterization of a Populus trichocarpa GATA TF, PtrGATA12, which encodes a nuclear-localized transcriptional activator predominantly expressing in developing xylem tissues. Overexpression of PtrGATA12 not only inhibited growths of most phenotypic traits and biomass accumulation, but also altered the expressions of some master TFs and pathway genes involved in secondary cell wall (SCW) and programmed cell death, leading to alternated SCW components and breaking forces of stems of transgenic lines. The significant changes occurred in the contents of hemicellulose and lignin and SCW thicknesses of fiber and vessel that increased by 13.5 and 10.8%, and 20.83 and 11.83%, respectively. Furthermore, PtrGATA12 bound directly to the promoters of a battery of TFs and pathway genes and activated them; the binding sites include two cis-acting elements that were specifically enriched in their promoter regions. Taken together, our results suggest PtrGATA12, as a higher hierarchical TF on the top of PtrWND6A, PtrWND6B, PtrMYB152, and PtrMYB21, exert a coordinated regulation of SCW components biosynthesis pathways through directly and indirectly controlling master TFs, middle-level TFs, and further downstream pathway genes of the currently known hierarchical transcription network that governs SCW formation.
Keywords: Populus trichocarpa; PtrGATA12; biological characterization; coordinated regulation; hierarchical regulator; secondary cell wall.
Copyright © 2021 Ren, Zhang, Liu, Liu, Tian, Cheng, Zhang, Wei and Wei.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
PtrHAT22, as a higher hierarchy regulator, coordinately regulates secondary cell wall component biosynthesis in Populus trichocarpa.Plant Sci. 2022 Mar;316:111170. doi: 10.1016/j.plantsci.2021.111170. Epub 2021 Dec 30. Plant Sci. 2022. PMID: 35151454
-
PtrWOX13A Promotes Wood Formation and Bioactive Gibberellins Biosynthesis in Populus trichocarpa.Front Plant Sci. 2022 Jun 28;13:835035. doi: 10.3389/fpls.2022.835035. eCollection 2022. Front Plant Sci. 2022. PMID: 35837467 Free PMC article.
-
Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa.Planta. 2017 May;245(5):927-938. doi: 10.1007/s00425-016-2640-1. Epub 2017 Jan 12. Planta. 2017. PMID: 28083709
-
Xylan in the Middle: Understanding Xylan Biosynthesis and Its Metabolic Dependencies Toward Improving Wood Fiber for Industrial Processing.Front Plant Sci. 2019 Feb 25;10:176. doi: 10.3389/fpls.2019.00176. eCollection 2019. Front Plant Sci. 2019. PMID: 30858858 Free PMC article. Review.
-
Recent Advances in the Transcriptional Regulation of Secondary Cell Wall Biosynthesis in the Woody Plants.Front Plant Sci. 2018 Oct 23;9:1535. doi: 10.3389/fpls.2018.01535. eCollection 2018. Front Plant Sci. 2018. PMID: 30405670 Free PMC article. Review.
Cited by
-
Comprehensive Transcriptome Analysis of Stem-Differentiating Xylem Upon Compression Stress in Cunninghamia Lanceolata.Front Genet. 2022 Mar 3;13:843269. doi: 10.3389/fgene.2022.843269. eCollection 2022. Front Genet. 2022. PMID: 35309135 Free PMC article.
-
Full-Length Transcriptome Sequencing and Comparative Transcriptomic Analyses Provide Comprehensive Insight Into Molecular Mechanisms of Cellulose and Lignin Biosynthesis in Cunninghamia lanceolata.Front Plant Sci. 2022 May 31;13:883720. doi: 10.3389/fpls.2022.883720. eCollection 2022. Front Plant Sci. 2022. PMID: 35712576 Free PMC article.
-
Construction of a genetic map and QTL mapping of seed size traits in soybean.Front Genet. 2023 Aug 25;14:1248315. doi: 10.3389/fgene.2023.1248315. eCollection 2023. Front Genet. 2023. PMID: 37693311 Free PMC article.
-
Deciphering the intricate hierarchical gene regulatory network: unraveling multi-level regulation and modifications driving secondary cell wall formation.Hortic Res. 2023 Dec 19;11(2):uhad281. doi: 10.1093/hr/uhad281. eCollection 2024 Feb. Hortic Res. 2023. PMID: 38344650 Free PMC article.
-
Spatiotemporal miRNA and transcriptomic network dynamically regulate the developmental and senescence processes of poplar leaves.Hortic Res. 2023 Sep 26;10(10):uhad186. doi: 10.1093/hr/uhad186. eCollection 2023 Oct. Hortic Res. 2023. PMID: 37899951 Free PMC article.
References
-
- An Y., Han X., Tang S., Xia X., Yin W. (2014). Poplar GATA transcription factor PdGNC is capable of regulating chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis under varying nitrogen levels. Plant Cell Tissue Organ Cult. 119, 313–327. 10.1007/s11240-014-0536-y - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

