Microarray analysis of the time-dependent expression profiles of long non-coding RNAs in the progression of vein graft stenotic disease
- PMID: 33968166
- PMCID: PMC8097238
- DOI: 10.3892/etm.2021.10067
Microarray analysis of the time-dependent expression profiles of long non-coding RNAs in the progression of vein graft stenotic disease
Abstract
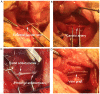
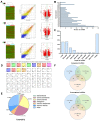
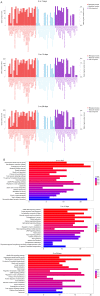
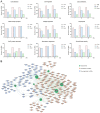
Long non-coding RNAs (lncRNAs) have been reported to be involved in various biological processes, including cell proliferation and apoptosis. However, the expression profiles of lncRNAs in patients with vein graft restenosis remain unknown. In the present study, the time-dependent expression profiles of genes in vein bypass grafting models were examined by microarray analysis. A total of 2,572 lncRNAs and 1,652 mRNAs were identified to be persistently significantly differentially expressed. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analysis was performed to investigate the functions of these lncRNAs. A total of 360 lncRNAs and 135 protein-coding genes were predicted to be involved in the vascular remodeling process. Co-expression network analysis revealed the association between 194 lncRNAs and seven associated protein-coding genes, including transforming growth factor-β1, Fes, Yes1 associated transcriptional regulator, sphingosine-1-phosphate receptor 1, Src, insulin receptor and melanoma cell adhesion molecule. Moreover, reverse transcription-quantitative PCR results supported those of the microarray data, and overexpression of AF062402, which regulates the transcription of Src, stimulated the proliferation of primary vascular smooth muscle cells. The findings of the present study may facilitate the development of novel therapeutic targets for vein graft restenosis and may help to improve the prognosis of patients following coronary artery bypass grafting.
Keywords: co-expression network; coronary artery bypass grafting; long non-coding RNAs; mRNAs; microarray analysis; vein graft stenotic disease.
Copyright: © Shen et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
