Transcriptomic analysis and competing endogenous RNA network in the human endometrium between proliferative and mid-secretory phases
- PMID: 33968190
- PMCID: PMC8097233
- DOI: 10.3892/etm.2021.10092
Transcriptomic analysis and competing endogenous RNA network in the human endometrium between proliferative and mid-secretory phases
Abstract
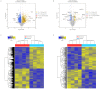
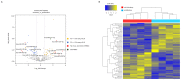
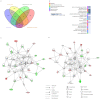
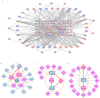
Successful embryo implantation is the first step for establishing natural pregnancy and is dependent on the crosstalk between the embryo and a receptive endometrium. However, the molecular signaling events for successful embryo implantation are not entirely understood. To identify differentially expressed transcripts [long-noncoding RNAs (lncRNAs), microRNAs (miRNAs) and mRNAs] and competing endogenous RNA (ceRNA) networks associated with endometrial receptivity, the current study analyzed gene expression profiles between proliferative and mid-secretory endometria in fertile women. A total of 247 lncRNAs, 67 miRNAs and 2,154 mRNAs were identified as differentially expressed between proliferative and mid-secretory endometria. Kyoto Encyclopedia of Genes and Genomes pathway analysis indicated that these differentially expressed genes were significantly enriched for 'cell adhesion molecules.' Additionally, 98 common mRNAs were significantly involved in tryptophan metabolism, metabolic pathways and FoxO signaling. From the differentially expressed lncRNA/miRNA/mRNA ceRNA network, hub RNAs that formed three axes were identified: The DLX6-AS1/miR-141 or miR-200a/OLFM1 axis, the WDFY3-AS2/miR-135a or miR-183/STC1 axis, and the LINC00240/miR-182/NDRG1 axis. These may serve important roles in the regulation of endometrial receptivity. The hub network of the current study may be developed as a candidate marker for endometrial receptivity.
Keywords: RNA sequencing; competing endogenous RNA network; endometrial receptivity; mid-secretory endometrium; proliferative endometrium.
Copyright: © Yu et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Genome-wide analysis of long noncoding RNAs, microRNAs, and mRNAs forming a competing endogenous RNA network in repeated implantation failure.Gene. 2019 Dec 15;720:144056. doi: 10.1016/j.gene.2019.144056. Epub 2019 Aug 19. Gene. 2019. PMID: 31437466
-
Construction of implantation failure related lncRNA-mRNA network and identification of lncRNA biomarkers for predicting endometrial receptivity.Int J Biol Sci. 2018 Jul 27;14(10):1361-1377. doi: 10.7150/ijbs.25081. eCollection 2018. Int J Biol Sci. 2018. PMID: 30123082 Free PMC article.
-
Integrated analysis of long noncoding RNA-associated competing endogenous RNA network in periodontitis.J Periodontal Res. 2018 Aug;53(4):495-505. doi: 10.1111/jre.12539. Epub 2018 Mar 8. J Periodontal Res. 2018. PMID: 29516510
-
Uterine fluid microRNAs are dysregulated in women with recurrent implantation failure.Hum Reprod. 2022 Apr 1;37(4):734-746. doi: 10.1093/humrep/deac019. Hum Reprod. 2022. PMID: 35147192 Free PMC article.
-
Differential expression of miRNAs and their target mRNAs in endometria prior to maternal recognition of pregnancy associates with endometrial receptivity for in vivo- and in vitro-produced bovine embryos.Biol Reprod. 2014 Dec;91(6):135. doi: 10.1095/biolreprod.114.121392. Epub 2014 Sep 24. Biol Reprod. 2014. PMID: 25253731
Cited by
-
Epigenetics of pregnancy: looking beyond the DNA code.J Assist Reprod Genet. 2022 Apr;39(4):801-816. doi: 10.1007/s10815-022-02451-x. Epub 2022 Mar 17. J Assist Reprod Genet. 2022. PMID: 35301622 Free PMC article. Review.
-
ATOH8 Expression Is Regulated by BMP2 and Plays a Key Role in Human Endometrial Stromal Cell Decidualization.Endocrinology. 2023 Nov 20;165(1):bqad188. doi: 10.1210/endocr/bqad188. Endocrinology. 2023. PMID: 38060684 Free PMC article.
-
Stanniocalcin Protein Expression in Female Reproductive Organs: Literature Review and Public Cancer Database Analysis.Endocrinology. 2024 Aug 27;165(10):bqae110. doi: 10.1210/endocr/bqae110. Endocrinology. 2024. PMID: 39186548 Free PMC article. Review.
-
Generating human blastoids modeling blastocyst-stage embryos and implantation.Nat Protoc. 2023 May;18(5):1584-1620. doi: 10.1038/s41596-023-00802-1. Epub 2023 Feb 15. Nat Protoc. 2023. PMID: 36792779 Free PMC article. Review.
-
Downregulated INHBB in endometrial tissue of recurrent implantation failure patients impeded decidualization through the ADCY1/cAMP signalling pathway.J Assist Reprod Genet. 2023 May;40(5):1135-1146. doi: 10.1007/s10815-023-02762-7. Epub 2023 Mar 13. J Assist Reprod Genet. 2023. PMID: 36913138 Free PMC article.
References
-
- Carson DD, Lagow E, Thathiah A, Al-Shami R, Farach-Carson MC, Vernon M, Yuan L, Fritz MA, Lessey B. Changes in gene expression during the early to mid-luteal (receptive phase) transition in human endometrium detected by high-density microarray screening. Mol Hum Reprod. 2002;8:871–879. doi: 10.1093/molehr/8.9.871. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
