Development and Validation of a Prognostic Gene Signature in Clear Cell Renal Cell Carcinoma
- PMID: 33968978
- PMCID: PMC8098777
- DOI: 10.3389/fmolb.2021.609865
Development and Validation of a Prognostic Gene Signature in Clear Cell Renal Cell Carcinoma
Abstract
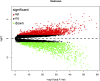
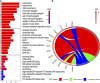
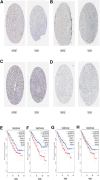
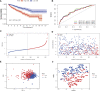
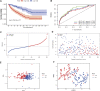
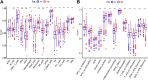
Clear cell renal cell carcinoma (ccRCC), one of the most common urologic cancer types, has a relatively good prognosis. However, clinical diagnoses are mostly done during the medium or late stages, when mortality and recurrence rates are quite high. Therefore, it is important to perform real-time information tracking and dynamic prognosis analysis for these patients. We downloaded the RNA-seq data and corresponding clinical information of ccRCC from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. A total of 3,238 differentially expressed genes were identified between normal and ccRCC tissues. Through a series of Weighted Gene Co-expression Network, overall survival, immunohistochemical and the least absolute shrinkage selection operator (LASSO) analyses, seven prognosis-associated genes (AURKB, FOXM1, PTTG1, TOP2A, TACC3, CCNA2, and MELK) were screened. Their risk score signature was then constructed. Survival analysis showed that high-risk scores exhibited significantly worse overall survival outcomes than low-risk patients. Accuracy of this prognostic signature was confirmed by the receiver operating characteristic curve and was further validated using another cohort. Gene set enrichment analysis showed that some cancer-associated phenotypes were significantly prevalent in the high-risk group. Overall, these findings prove that this risk model can potentially improve individualized diagnostic and therapeutic strategies.
Keywords: WGCNA; kidney cancer; microarray; novel markers; prognostic model; targeting therapy.
Copyright © 2021 Zhan, Wang, Xu, Huang, Su, Chen, Wang, Qi and Bai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

