Alpha 1 Antitrypsin is an Inhibitor of the SARS-CoV-2-Priming Protease TMPRSS2
- PMID: 33969249
- PMCID: PMC8097828
- DOI: 10.20411/pai.v6i1.408
Alpha 1 Antitrypsin is an Inhibitor of the SARS-CoV-2-Priming Protease TMPRSS2
Abstract
Background: Host proteases have been suggested to be crucial for dissemination of MERS, SARS-CoV, and SARS-CoV-2 coronaviruses, but the relative contribution of membrane versus intracellular proteases remains controversial. Transmembrane serine protease 2 (TMPRSS2) is regarded as one of the main proteases implicated in the coronavirus S protein priming, an important step for binding of the S protein to the angiotensin-converting enzyme 2 (ACE2) receptor before cell entry.
Methods: We developed a cell-based assay to identify TMPRSS2 inhibitors. Inhibitory activity was established in SARS-CoV-2 viral load systems.
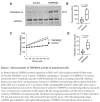
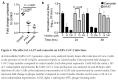
Results: We identified the human extracellular serine protease inhibitor (serpin) alpha 1 anti-trypsin (A1AT) as a novel TMPRSS2 inhibitor. Structural modeling revealed that A1AT docked to an extracellular domain of TMPRSS2 in a conformation that is suitable for catalysis, resembling similar serine protease inhibitor complexes. Inhibitory activity of A1AT was established in a SARS-CoV-2 viral load system. Notably, plasma A1AT levels were associated with COVID-19 disease severity.
Conclusions: Our data support the key role of extracellular serine proteases in SARS CoV-2 infections and indicate that treatment with serpins, particularly the FDA-approved drug A1AT, may be effective in limiting SARS-CoV-2 dissemination by affecting the surface of the host cells.
Keywords: COVID; TMPRSS2; alpha 1 antitrypsin; camostat mesylate; coronavirus; protease.
Copyright © Pathogens and Immunity 2021.
Conflict of interest statement
M.E.R. is a consultant for Pulm One, Spoon Guru, ClostraBio, Serpin Pharma, Celgene, Astra Zeneca, Allakos, Arena Pharmaceuticals, Guidepoint, and Suvretta Capital Management and has an equity interest in the first 4 listed and royalties from reslizumab (Teva Pharmaceuticals), PEESSv2 (Mapi Research Trust), and UpToDate. M.E.R. is an inventor of patents owned by Cincinnati Children's Hospital. M.E.R. and N.P.A. are inventors of a patent owned by Cincinnati Children's Hospital with the provisional number of 63/017,027.
Figures



Update of
-
Alpha 1 Antitrypsin is an Inhibitor of the SARS-CoV-2-Priming Protease TMPRSS2.bioRxiv [Preprint]. 2020 Oct 7:2020.05.04.077826. doi: 10.1101/2020.05.04.077826. bioRxiv. 2020. Update in: Pathog Immun. 2021 Apr 26;6(1):55-74. doi: 10.20411/pai.v6i1.408. PMID: 33052338 Free PMC article. Updated. Preprint.
References
-
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80 e8. doi: 10.1016/j.cell.2020.02.052. PubMed PMID: ; PMCID: . - DOI - PMC - PubMed
-
- Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88(2):1293–307. doi: 10.1128/JVI.02202-13. PubMed PMID: ; PMCID: . - DOI - PMC - PubMed
-
- Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, Nagata N, Sekizuka T, Katoh H, Kato F, Sakata M, Tahara M, Kutsuna S, Ohmagari N, Kuroda M, Suzuki T, Kageyama T, Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117(13):7001–3. doi: 10.1073/pnas.2002589117. PubMed PMID: ; PMCID: . - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
