Histone H3 N-terminal mimicry drives a novel network of methyl-effector interactions
- PMID: 33969871
- PMCID: PMC8166343
- DOI: 10.1042/BCJ20210203
Histone H3 N-terminal mimicry drives a novel network of methyl-effector interactions
Abstract
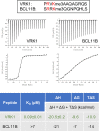
The reader ability of PHD fingers is largely limited to the recognition of the histone H3 N-terminal tail. Distinct subsets of PHDs bind either H3K4me3 (a transcriptional activator mark) or H3K4me0 (a transcriptional repressor state). Structural studies have identified common features among the different H3K4me3 effector PHDs, including (1) removal of the initiator methionine residue of H3 to prevent steric interference, (2) a groove where arginine-2 binds, and (3) an aromatic cage that engages methylated lysine-4. We hypothesize that some PHDs might have the ability to engage with non-histone ligands, as long as they adhere to these three rules. A search of the human proteome revealed an enrichment of chromatin-binding proteins that met these criteria, which we termed H3 N-terminal mimicry proteins (H3TMs). Seven H3TMs were selected, and used to screen a protein domain microarray for potential effector domains, and they all had the ability to bind H3K4me3-interacting effector domains. Furthermore, the binding affinity between the VRK1 peptide and the PHD domain of PHF2 is ∼3-fold stronger than that of PHF2 and H3K4me3 interaction. The crystal structure of PHF2 PHD finger bound with VRK1 K4me3 peptide provides a molecular basis for stronger binding of VRK1 peptide. In addition, a number of the H3TMs peptides, in their unmethylated form, interact with NuRD transcriptional repressor complex. Our findings provide in vitro evidence that methylation of H3TMs can promote interactions with PHD and Tudor domain-containing proteins and potentially block interactions with the NuRD complex. We propose that these interactions can occur in vivo as well.
Keywords: NuRD complex; PHD domain; PHF2; Tudor domain; VRK1.
© 2021 The Author(s).
Conflict of interest statement
M.T.B. is a co-founder of EpiCypher.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

