The order of PDZ3 and TrpCage in fusion chimeras determines their properties-a biophysical characterization
- PMID: 33969912
- PMCID: PMC8284584
- DOI: 10.1002/pro.4107
The order of PDZ3 and TrpCage in fusion chimeras determines their properties-a biophysical characterization
Abstract
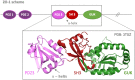
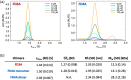
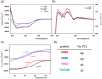
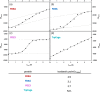
Most of the structural proteins known today are composed of domains that carry their own functions while keeping their structural properties. It is supposed that such domains, when taken out of the context of the whole protein, can retain their original structure and function to a certain extent. Information on the specific functional and structural characteristics of individual domains in a new context of artificial fusion proteins may help to reveal the rules of internal and external domain communication. Moreover, this could also help explain the mechanism of such communication and address how the mutual allosteric effect plays a role in a such multi-domain protein system. The simple model system of the two-domain fusion protein investigated in this work consisted of a well-folded PDZ3 domain and an artificially designed small protein domain called Tryptophan Cage (TrpCage). Two fusion proteins with swapped domain order were designed to study their structural and functional features as well as their biophysical properties. The proteins composed of PDZ3 and TrpCage, both identical in amino acid sequence but different in composition (PDZ3-TrpCage, TrpCage-PDZ3), were studied using circualr dichroism (CD) spectrometry, analytical ultracentrifugation, and molecular dynamic simulations. The biophysical analysis uncovered different structural and denaturation properties of both studied proteins, revealing their different unfolding pathways and dynamics.
Keywords: chimeras; fusion protein; protein domains; protein dynamic studies.
© 2021 The Protein Society.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

