Knowledge bases and software support for variant interpretation in precision oncology
- PMID: 33971666
- PMCID: PMC8574624
- DOI: 10.1093/bib/bbab134
Knowledge bases and software support for variant interpretation in precision oncology
Erratum in
-
Knowledge bases and software support for variant interpretation in precision oncology.Brief Bioinform. 2021 Nov 5;22(6):bbab246. doi: 10.1093/bib/bbab246. Brief Bioinform. 2021. PMID: 34125166 Free PMC article. No abstract available.
Abstract
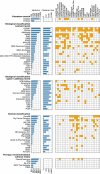
Precision oncology is a rapidly evolving interdisciplinary medical specialty. Comprehensive cancer panels are becoming increasingly available at pathology departments worldwide, creating the urgent need for scalable cancer variant annotation and molecularly informed treatment recommendations. A wealth of mainly academia-driven knowledge bases calls for software tools supporting the multi-step diagnostic process. We derive a comprehensive list of knowledge bases relevant for variant interpretation by a review of existing literature followed by a survey among medical experts from university hospitals in Germany. In addition, we review cancer variant interpretation tools, which integrate multiple knowledge bases. We categorize the knowledge bases along the diagnostic process in precision oncology and analyze programmatic access options as well as the integration of knowledge bases into software tools. The most commonly used knowledge bases provide good programmatic access options and have been integrated into a range of software tools. For the wider set of knowledge bases, access options vary across different parts of the diagnostic process. Programmatic access is limited for information regarding clinical classifications of variants and for therapy recommendations. The main issue for databases used for biological classification of pathogenic variants and pathway context information is the lack of standardized interfaces. There is no single cancer variant interpretation tool that integrates all identified knowledge bases. Specialized tools are available and need to be further developed for different steps in the diagnostic process.
Keywords: HiGHmed; cancer therapy; data integration; molecular tumor board; personalized medicine.
© The Author(s) 2021. Published by Oxford University Press.
Figures

References
-
- Horak P, Klink B, Heining C, et al. Precision oncology based on omics data: the NCT Heidelberg experience. Int J Cancer 2017. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

