Adaptive laboratory evolution accelerated glutarate production by Corynebacterium glutamicum
- PMID: 33971881
- PMCID: PMC8112011
- DOI: 10.1186/s12934-021-01586-3
Adaptive laboratory evolution accelerated glutarate production by Corynebacterium glutamicum
Abstract
Background: The demand for biobased polymers is increasing steadily worldwide. Microbial hosts for production of their monomeric precursors such as glutarate are developed. To meet the market demand, production hosts have to be improved constantly with respect to product titers and yields, but also shortening bioprocess duration is important.
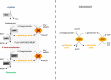
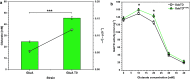
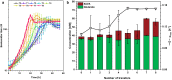
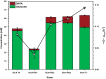
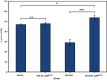
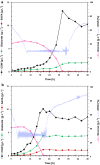
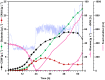
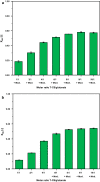
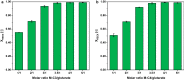
Results: In this study, adaptive laboratory evolution was used to improve a C. glutamicum strain engineered for production of the C5-dicarboxylic acid glutarate by flux enforcement. Deletion of the L-glutamic acid dehydrogenase gene gdh coupled growth to glutarate production since two transaminases in the glutarate pathway are crucial for nitrogen assimilation. The hypothesis that strains selected for faster glutarate-coupled growth by adaptive laboratory evolution show improved glutarate production was tested. A serial dilution growth experiment allowed isolating faster growing mutants with growth rates increasing from 0.10 h-1 by the parental strain to 0.17 h-1 by the fastest mutant. Indeed, the fastest growing mutant produced glutarate with a twofold higher volumetric productivity of 0.18 g L-1 h-1 than the parental strain. Genome sequencing of the evolved strain revealed candidate mutations for improved production. Reverse genetic engineering revealed that an amino acid exchange in the large subunit of L-glutamic acid-2-oxoglutarate aminotransferase was causal for accelerated glutarate production and its beneficial effect was dependent on flux enforcement due to deletion of gdh. Performance of the evolved mutant was stable at the 2 L bioreactor-scale operated in batch and fed-batch mode in a mineral salts medium and reached a titer of 22.7 g L-1, a yield of 0.23 g g-1 and a volumetric productivity of 0.35 g L-1 h-1. Reactive extraction of glutarate directly from the fermentation broth was optimized leading to yields of 58% and 99% in the reactive extraction and reactive re-extraction step, respectively. The fermentation medium was adapted according to the downstream processing results.
Conclusion: Flux enforcement to couple growth to operation of a product biosynthesis pathway provides a basis to select strains growing and producing faster by adaptive laboratory evolution. After identifying candidate mutations by genome sequencing causal mutations can be identified by reverse genetics. As exemplified here for glutarate production by C. glutamicum, this approach allowed deducing rational metabolic engineering strategies.
Keywords: Adaptive laboratory evolution; Corynebacterium glutamicum; Glutarate; Metabolic engineering; Reactive extraction; Reverse genetics; Volumetric productivity.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Systems metabolic engineering of Corynebacterium glutamicum for the production of the carbon-5 platform chemicals 5-aminovalerate and glutarate.Microb Cell Fact. 2016 Sep 13;15(1):154. doi: 10.1186/s12934-016-0553-0. Microb Cell Fact. 2016. PMID: 27618862 Free PMC article.
-
Systems metabolic engineering of Corynebacterium glutamicum eliminates all by-products for selective and high-yield production of the platform chemical 5-aminovalerate.Metab Eng. 2022 Sep;73:168-181. doi: 10.1016/j.ymben.2022.07.005. Epub 2022 Jul 30. Metab Eng. 2022. PMID: 35917915
-
Metabolic engineering of Corynebacterium glutamicum for enhanced production of 5-aminovaleric acid.Microb Cell Fact. 2016 Oct 7;15(1):174. doi: 10.1186/s12934-016-0566-8. Microb Cell Fact. 2016. PMID: 27717386 Free PMC article.
-
Boosting Anaplerotic Reactions by Pyruvate Kinase Gene Deletion and Phosphoenolpyruvate Carboxylase Desensitization for Glutamic Acid and Lysine Production in Corynebacterium glutamicum.Adv Biochem Eng Biotechnol. 2017;159:181-198. doi: 10.1007/10_2016_31. Adv Biochem Eng Biotechnol. 2017. PMID: 27872961 Review.
-
Strategy for improving L-isoleucine production efficiency in Corynebacterium glutamicum.Appl Microbiol Biotechnol. 2019 Mar;103(5):2101-2111. doi: 10.1007/s00253-019-09632-2. Epub 2019 Jan 20. Appl Microbiol Biotechnol. 2019. PMID: 30663007 Review.
Cited by
-
Efficient cell factories for the production of N-methylated amino acids and for methanol-based amino acid production.Microb Biotechnol. 2022 Aug;15(8):2145-2159. doi: 10.1111/1751-7915.14067. Epub 2022 Apr 30. Microb Biotechnol. 2022. PMID: 35488805 Free PMC article. Review.
-
Metabolic Engineering of Corynebacterium glutamicum for Sustainable Production of the Aromatic Dicarboxylic Acid Dipicolinic Acid.Microorganisms. 2022 Mar 29;10(4):730. doi: 10.3390/microorganisms10040730. Microorganisms. 2022. PMID: 35456781 Free PMC article.
-
Intelligent host engineering for metabolic flux optimisation in biotechnology.Biochem J. 2021 Oct 29;478(20):3685-3721. doi: 10.1042/BCJ20210535. Biochem J. 2021. PMID: 34673920 Free PMC article. Review.
-
Production of indole by Corynebacterium glutamicum microbial cell factories for flavor and fragrance applications.Microb Cell Fact. 2022 Mar 24;21(1):45. doi: 10.1186/s12934-022-01771-y. Microb Cell Fact. 2022. PMID: 35331232 Free PMC article.
-
Utilization of a Wheat Sidestream for 5-Aminovalerate Production in Corynebacterium glutamicum.Front Bioeng Biotechnol. 2021 Sep 29;9:732271. doi: 10.3389/fbioe.2021.732271. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34660554 Free PMC article.
References
-
- European Bioplastics e.V. Bioplastics market data 2021. https://www.european-bioplastics.org/market/. Accessed 22 Feb 2021.
-
- Radzik P, Leszczyńska A, Pielichowski K. Modern biopolyamide-based materials: synthesis and modification. Polym Bull. 2020;77:501–528. doi: 10.1007/s00289-019-02718-x. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

