Identification of an immune classification for cervical cancer and integrative analysis of multiomics data
- PMID: 33971902
- PMCID: PMC8111986
- DOI: 10.1186/s12967-021-02845-y
Identification of an immune classification for cervical cancer and integrative analysis of multiomics data
Abstract
Background: To understand the molecular mechanisms of the antitumour response, we analysed the immune landscape of cervical cancer to identify novel immune molecular classes.
Methods: We established a stable immune molecular classification using a nonnegative matrix factorization algorithm and validated the correlation in two validation sets of 249 samples.
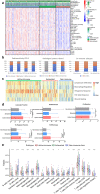
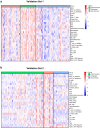
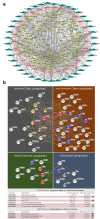
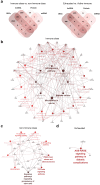
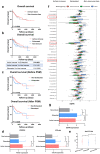
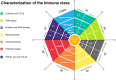
Results: Approximately 78% of cervical cancers (CCs) (228/293) were identified to show significant enrichment in immune cells (e.g., CD8 T cells and macrophages), a type I IFN response, enhanced cytolytic activity and high PDCD1, and these CCs were referred to as the "immune class". We further identified two subtypes of the immune class: active immune and exhausted subtypes. Although the active immune subtype was characterized by enrichment of IFN signatures and better survival, the exhausted subtype expressed activated stroma, a wound healing signature, enhanced M2 macrophages and absence of CD8 T cells and the TGF-β response signature. Integrative analysis of multiomics data identified EGFR, JUN, MYC, FN1 and SERPINE1 as key modulators of the tumour immune microenvironment and potential targets for combination therapies which was validated in two validation sets.
Conclusions: Our study introduces a novel immune classification that might predict ideal candidates to receive immunotherapy or specific combination therapies.
Keywords: Cervical cancer; Immune classification; Multiomics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: implications for immunotherapy.Ann Oncol. 2019 Jan 1;30(1):68-75. doi: 10.1093/annonc/mdy470. Ann Oncol. 2019. PMID: 30407504
-
Identification and validation of tumour microenvironment-based immune molecular subgroups for gastric cancer: immunotherapeutic implications.Cancer Immunol Immunother. 2020 Jun;69(6):1057-1069. doi: 10.1007/s00262-020-02525-8. Epub 2020 Feb 25. Cancer Immunol Immunother. 2020. PMID: 32100076 Free PMC article.
-
The correlation between immune subtypes and consensus molecular subtypes in colorectal cancer identifies novel tumour microenvironment profiles, with prognostic and therapeutic implications.Eur J Cancer. 2019 Dec;123:118-129. doi: 10.1016/j.ejca.2019.09.008. Epub 2019 Nov 1. Eur J Cancer. 2019. PMID: 31678770
-
Efficacy Against Human Prostate Cancer by Prostate-specific Membrane Antigen-specific, Transforming Growth Factor-β Insensitive Genetically Targeted CD8+ T-cells Derived from Patients with Metastatic Castrate-resistant Disease.Eur Urol. 2018 May;73(5):648-652. doi: 10.1016/j.eururo.2017.12.008. Epub 2017 Dec 21. Eur Urol. 2018. PMID: 29275833 Free PMC article. Review.
-
Blockade of TGF-β signaling: a potential target for cancer immunotherapy?Expert Opin Ther Targets. 2019 Aug;23(8):679-693. doi: 10.1080/14728222.2019.1636034. Epub 2019 Jun 27. Expert Opin Ther Targets. 2019. PMID: 31232607 Review.
Cited by
-
Multi-omics analyses construct an inflammatory response based prognostic gene signature for cervical cancer and suggest tumor infiltrating monocytes subgroups as key players.Front Immunol. 2025 May 19;16:1563593. doi: 10.3389/fimmu.2025.1563593. eCollection 2025. Front Immunol. 2025. PMID: 40458391 Free PMC article.
-
Sequencing-based transcriptome analysis reveals diversification of immune response- and angiogenesis-related expression patterns of early-stage cervical carcinoma as compared with high-grade CIN.Front Immunol. 2023 Sep 4;14:1215607. doi: 10.3389/fimmu.2023.1215607. eCollection 2023. Front Immunol. 2023. PMID: 37731500 Free PMC article.
-
Integrative analysis of gene expression and DNA methylation to identify biomarkers of non-genital warts induced by low-risk human papillomaviruses infection.Heliyon. 2023 May 7;9(5):e16101. doi: 10.1016/j.heliyon.2023.e16101. eCollection 2023 May. Heliyon. 2023. PMID: 37215908 Free PMC article.
-
Netie: inferring the evolution of neoantigen-T cell interactions in tumors.Nat Methods. 2022 Nov;19(11):1480-1489. doi: 10.1038/s41592-022-01644-7. Epub 2022 Oct 27. Nat Methods. 2022. PMID: 36303017 Free PMC article.
-
T cell subsets in cervical cancer tumor microenvironment: advances and therapeutic opportunities.Front Immunol. 2025 Jun 5;16:1612032. doi: 10.3389/fimmu.2025.1612032. eCollection 2025. Front Immunol. 2025. PMID: 40539072 Free PMC article. Review.
References
-
- Chung HC, Ros W, Delord JP, Perets R, Italiano A, Shapira-Frommer R, Manzuk L, Piha-Paul SA, Xu L, Zeigenfuss S, et al. Efficacy and safety of pembrolizumab in previously treated advanced cervical cancer: results from the phase II KEYNOTE-158 Study. J Clin Oncol. 2019;37:1470–1478. doi: 10.1200/JCO.18.01265. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

