Cryopreservation of human cancers conserves tumour heterogeneity for single-cell multi-omics analysis
- PMID: 33971952
- PMCID: PMC8111910
- DOI: 10.1186/s13073-021-00885-z
Cryopreservation of human cancers conserves tumour heterogeneity for single-cell multi-omics analysis
Abstract
Background: High throughput single-cell RNA sequencing (scRNA-Seq) has emerged as a powerful tool for exploring cellular heterogeneity among complex human cancers. scRNA-Seq studies using fresh human surgical tissue are logistically difficult, preclude histopathological triage of samples, and limit the ability to perform batch processing. This hindrance can often introduce technical biases when integrating patient datasets and increase experimental costs. Although tissue preservation methods have been previously explored to address such issues, it is yet to be examined on complex human tissues, such as solid cancers and on high throughput scRNA-Seq platforms.
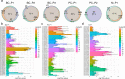
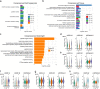
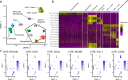
Methods: Using the Chromium 10X platform, we sequenced a total of ~ 120,000 cells from fresh and cryopreserved replicates across three primary breast cancers, two primary prostate cancers and a cutaneous melanoma. We performed detailed analyses between cells from each condition to assess the effects of cryopreservation on cellular heterogeneity, cell quality, clustering and the identification of gene ontologies. In addition, we performed single-cell immunophenotyping using CITE-Seq on a single breast cancer sample cryopreserved as solid tissue fragments.
Results: Tumour heterogeneity identified from fresh tissues was largely conserved in cryopreserved replicates. We show that sequencing of single cells prepared from cryopreserved tissue fragments or from cryopreserved cell suspensions is comparable to sequenced cells prepared from fresh tissue, with cryopreserved cell suspensions displaying higher correlations with fresh tissue in gene expression. We showed that cryopreservation had minimal impacts on the results of downstream analyses such as biological pathway enrichment. For some tumours, cryopreservation modestly increased cell stress signatures compared to freshly analysed tissue. Further, we demonstrate the advantage of cryopreserving whole-cells for detecting cell-surface proteins using CITE-Seq, which is impossible using other preservation methods such as single nuclei-sequencing.
Conclusions: We show that the viable cryopreservation of human cancers provides high-quality single-cells for multi-omics analysis. Our study guides new experimental designs for tissue biobanking for future clinical single-cell RNA sequencing studies.
Keywords: Breast cancer; CITE-Seq; Cryopreservation; Melanoma; Prostate cancer; Single-cell RNA sequencing; Tumour heterogeneity.
Conflict of interest statement
No conflicts of interests. R.A.S. has received fees for professional services from Merck Sharp & Dohme, GlaxoSmithKline Australia, Bristol-Myers Squibb, Novartis, Myriad, NeraCare and Amgen. G.V.L. is consultant advisor to Aduro, BMS, Mass-Array, Merck MSD, Novartis, Pierre-Fabre, Roche, Sandoz and Qbiotics. The remaining authors declare that they have no competing interests.
Figures



References
-
- Madissoon E, Wilbrey-Clark A, Miragaia RJ, Saeb-Parsy K, Mahbubani KT, Georgakopoulos N, Harding P, Polanski K, Huang N, Nowicki-Osuch K, et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 2019;21:1. doi: 10.1186/s13059-019-1906-x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

