DHODH and cancer: promising prospects to be explored
- PMID: 33971967
- PMCID: PMC8107416
- DOI: 10.1186/s40170-021-00250-z
DHODH and cancer: promising prospects to be explored
Abstract
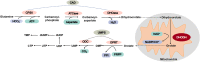
Human dihydroorotate dehydrogenase (DHODH) is a flavin-dependent mitochondrial enzyme catalyzing the fourth step in the de novo pyrimidine synthesis pathway. It is originally a target for the treatment of the non-neoplastic diseases involving in rheumatoid arthritis and multiple sclerosis, and is re-emerging as a validated therapeutic target for cancer therapy. In this review, we mainly unravel the biological function of DHODH in tumor progression, including its crucial role in de novo pyrimidine synthesis and mitochondrial respiratory chain in cancer cells. Moreover, various DHODH inhibitors developing in the past decades are also been displayed, and the specific mechanism between DHODH and its additional effects are illustrated. Collectively, we detailly discuss the association between DHODH and tumors in recent years here, and believe it will provide significant evidences and potential strategies for utilizing DHODH as a potential target in preclinical and clinical cancer therapies.
Keywords: Cancer metabolism; DHODH inhibitors; De novo pyrimidine biosynthesis; Dihydroorotate dehydrogenase; Mitochondria.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Barnes T, Parry P, Hart I, Jones C, Minet M, Patterson D. Regional mapping of the gene encoding dihydroorotate dehydrogenase, an enzyme involved in UMP synthesis, electron transport, and superoxide generation, to human chromosome region 16q22. Somat Cell Mol Genet. 1993;19:405–411. doi: 10.1007/BF01232751. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

