RNase P Inhibitors Identified as Aggregators
- PMID: 33972249
- PMCID: PMC8284443
- DOI: 10.1128/AAC.00300-21
RNase P Inhibitors Identified as Aggregators
Abstract
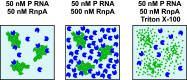
RNase P is an essential enzyme responsible for tRNA 5'-end maturation. In most bacteria, the enzyme is a ribonucleoprotein consisting of a catalytic RNA subunit and a small protein cofactor termed RnpA. Several studies have reported small-molecule inhibitors directed against bacterial RNase P that were identified by high-throughput screenings. Using the bacterial RNase P enzymes from Thermotoga maritima, Bacillus subtilis, and Staphylococcus aureus as model systems, we found that such compounds, including RNPA2000 (and its derivatives), iriginol hexaacetate, and purpurin, induce the formation of insoluble aggregates of RnpA rather than acting as specific inhibitors. In the case of RNPA2000, aggregation was induced by Mg2+ ions. These findings were deduced from solubility analyses by microscopy and high-performance liquid chromatography (HPLC), RnpA-inhibitor co-pulldown experiments, detergent addition, and RnpA titrations in enzyme activity assays. Finally, we used a B. subtilis RNase P depletion strain, whose lethal phenotype could be rescued by a protein-only RNase P of plant origin, for inhibition zone analyses on agar plates. These cell-based experiments argued against RNase P-specific inhibition of bacterial growth by RNPA2000. We were also unable to confirm the previously reported nonspecific RNase activity of S. aureus RnpA itself. Our results indicate that high-throughput screenings searching for bacterial RNase P inhibitors are prone to the identification of "false positives" that are also termed
Keywords: RNase P inhibitors; RnpA protein subunit; bacterial RNase P; protein aggregators.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

