Levels and Characteristics of mRNAs in Spores of Firmicute Species
- PMID: 33972352
- PMCID: PMC8315741
- DOI: 10.1128/JB.00017-21
Levels and Characteristics of mRNAs in Spores of Firmicute Species
Abstract
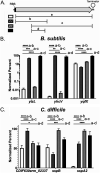
Spores of firmicute species contain 100s of mRNAs, whose major function in Bacillus subtilis is to provide ribonucleotides for new RNA synthesis when spores germinate. To determine if this is a general phenomenon, RNA was isolated from spores of multiple firmicute species and relative mRNA levels determined by transcriptome sequencing (RNA-seq). Determination of RNA levels in single spores allowed calculation of RNA nucleotides/spore, and assuming mRNA is 3% of spore RNA indicated that only ∼6% of spore mRNAs were present at >1/spore. Bacillus subtilis, Bacillus atrophaeus, and Clostridioides difficile spores had 49, 42, and 51 mRNAs at >1/spore, and numbers of mRNAs at ≥1/spore were ∼10 to 50% higher in Geobacillus stearothermophilus and Bacillus thuringiensis Al Hakam spores and ∼4-fold higher in Bacillus megaterium spores. In all species, some to many abundant spore mRNAs (i) were transcribed by RNA polymerase with forespore-specific σ factors, (ii) encoded proteins that were homologs of those encoded by abundant B. subtilis spore mRNAs and are proteins in dormant spores, and (iii) were likely transcribed in the mother cell compartment of the sporulating cell. Analysis of the coverage of RNA-seq reads on mRNAs from all species suggested that abundant spore mRNAs were fragmented, as was confirmed by reverse transcriptase quantitative PCR (RT-qPCR) analysis of abundant B. subtilis and C. difficile spore mRNAs. These data add to evidence indicating that the function of at least the great majority of mRNAs in all firmicute spores is to be degraded to generate ribonucleotides for new RNA synthesis when spores germinate. IMPORTANCE Only ∼6% of mRNAs in spores of six firmicute species are at ≥1 molecule/spore, many abundant spore mRNAs encode proteins similar to B. subtilis spore proteins, and some abundant B. subtilis and C. difficile spore mRNAs were fragmented. Most of the abundant B. subtilis and other Bacillales spore mRNAs are transcribed under the control of the forespore-specific RNA polymerase σ factors, F or G, and these results may stimulate transcription analyses in developing spores of species other than B. subtilis. These findings, plus the absence of key nucleotide biosynthetic enzymes in spores, suggest that firmicute spores' abundant mRNAs are not translated when spores germinate but instead are degraded to generate ribonucleotides for new RNA synthesis by the germinated spore.
Keywords: Bacillus subtilis; Bacillus thuringiensis; Clostridioides; Geobacillus; mRNA; spores.
Figures
References
-
- Setlow P. 2016. Spore resistance properties, p 201–215. In Eichenberger P, Driks A (ed), The bacterial spore: from molecules to systems. ASM Press, Washington, DC.
-
- Setlow P, Johnson EA. 2012. Spores and their significance, p 45–79. In Doyle MP, Buchanan R (ed), Food microbiology: fundamentals and frontiers, 4th ed. ASM Press, Washington, DC.
-
- Arrieta-Ortiz ML, Hafemeister C, Bate AR, Chu Greenfield TA, Shuster B, Barry SN, Gallitto M, Liu B, Kacmarczyk T, Santoriello F, Chen J, Rodrigues CD, Sato T, Rudner DZ, Driks A, Bonneau R, Eichenberger P. 2015. An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network. Mol Syst Biol 11:839. doi: 10.15252/msb.20156236. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

